If you find our code or paper helpful, please consider starring our repository and citing:
@article{pinyoanuntapong2024controlmm,
title={ControlMM: Controllable Masked Motion Generation},
author={Pinyoanuntapong, Ekkasit and Saleem, Muhammad Usama and Karunratanakul, Korrawe and Wang, Pu and Xue, Hongfei and Chen, Chen and Guo, Chuan and Cao, Junli and Ren, Jian and Tulyakov, Sergey},
journal={arXiv preprint arXiv:2410.10780},
year={2024}
}
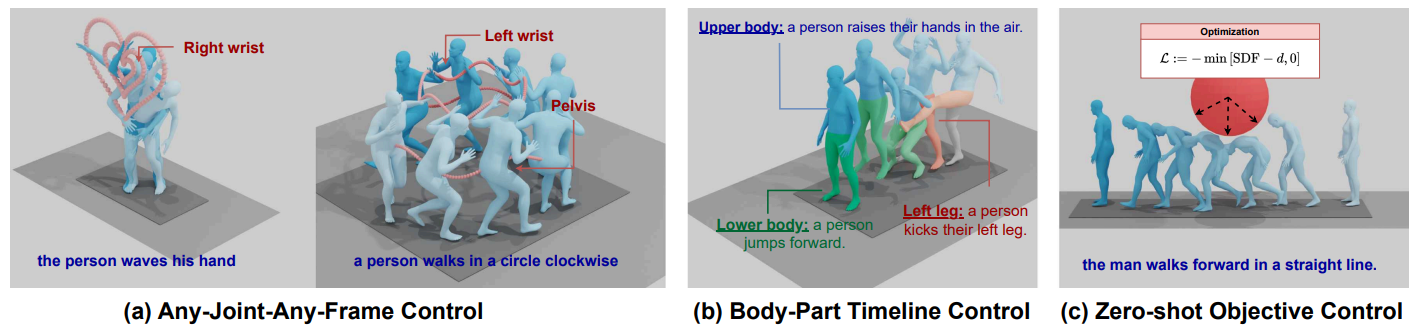
- Joint Control (GMD, OmniControl, and MMM Evaluation)
- ProMoGen Evaluation
- STMC Evaluation
- Joint Control
- Obstacle Avoidance
- Body Part Timeline Control
- Retrain MoMask with Cross Entropy for All Positions
- Add Logits Regularizer
Our code built on top of MoMask. If you encounter any issues, please refer to the MoMask repository for setup and troubleshooting instructions.
conda env create -f environment.yml
conda activate ControlMM
pip install git+https://github.com/openai/CLIP.git
pip install -r requirements.txt
bash prepare/download_models.sh
For evaluation only.
bash prepare/download_evaluator.sh
bash prepare/download_glove.sh
You have two options here:
- Skip getting data, if you just want to generate motions using own descriptions.
- Get full data, if you want to re-train and evaluate the model.
(a). Full data (text + motion)
HumanML3D - Follow the instruction in HumanML3D, then copy the result dataset to our repository:
cp -r ../HumanML3D/HumanML3D ./dataset/HumanML3D
python eval_t2m_trans_res.py \
--res_name tres_nlayer8_ld384_ff1024_rvq6ns_cdp0.2_sw \
--dataset_name t2m \
--ctrl_name 'z2024-08-23-01-27-51_CtrlNet_randCond1-196_l1.1XEnt.9TTT__fixRandCond' \
--gpu_id 0 \
--ext 0_each100Last600CtrnNet \
--control trajectory \
--density -1 \
--each_iter 100 \
--last_iter 600 \
--ctrl_net T
python eval_t2m_trans_res.py \
--res_name tres_nlayer8_ld384_ff1024_rvq6ns_cdp0.2_sw \
--dataset_name t2m \
--ctrl_name 'z2024-08-27-21-07-55_CtrlNet_randCond1-196_l1.5XEnt.5TTT__cross' \
--gpu_id 4 \
--ext 0_each100_last600_ctrlNetT \
--control cross \
--density -1 \
--each_iter 100 \
--last_iter 600 \
--ctrl_net T
The following joints can be controlled:
[pelvis, left_foot, right_foot, head, left_wrist, right_wrist]
| Argument | Description |
|---|---|
--res_name |
Name of the residual transformer |
--ctrl_name |
Name of the control transformer (VQ and Masked Transformer are also saved in this) |
--gpu_id |
GPU ID to use |
--ext |
Log name used for saving results, stored in: checkpoints/t2m/{ctrl_name}/eval/{ext} |
--control |
Type of random joint control: • trajectory – pelvis only• random – uniform random joints• cross – random combinations, see section [A.11 CROSS COMBINATION]• Any single joint: pelvis, l_foot, r_foot, head, left_wrist, right_wrist, lower• all – all joints |
--density |
Number of control frames: • 1, 2, 5 – exact number of control frames• 49 – 25% of ground truth length• 196 – 100% of ground truth length(If GT length < 196, 49/196 are converted proportionally) |
--each_iter |
Number of logits optimization iterations at each unmask step |
--last_iter |
Number of logits optimization iterations at the last unmask step |
--ctrl_net |
Enable ControlNet with Logits Regularizer: T or F |
python -m generation.control_joint --path_name ./output/control1 --iter_each 100 --iter_last 600
| Argument | Type | Default | Description |
|---|---|---|---|
--path_name |
str | ./output/test |
Output directory to save the optimization results. |
--iter_each |
int | 100 |
Number of logits optimization steps at each unmasking step. |
--iter_last |
int | 600 |
Number of logits optimization steps at the final unmasking step. |
--show |
flag | False |
If set, automatically opens the result HTML visualization after execution. |
We sincerely thank the open-sourcing of these works where our code is based on: MoMask, OmniControl, GMD, MMM, TLControl, STMC, ProgMoGen, TEMOS and BAMM
This code is distributed under an LICENSE-CC-BY-NC-ND-4.0.
Note that our code depends on other libraries, including SMPL, SMPL-X, PyTorch3D, and uses datasets which each have their own respective licenses that must also be followed.