Automated Community Detection of Cell Populations in Python
This repo contains the current Python implementation of ACDC, an optimization-based framework to automatize clustering of cell populations from scRNA-seq data using community detection algorithms.
acdc_py is deployed as a Python package and fully compatible with Scanpy.
Why you may find this package useful:
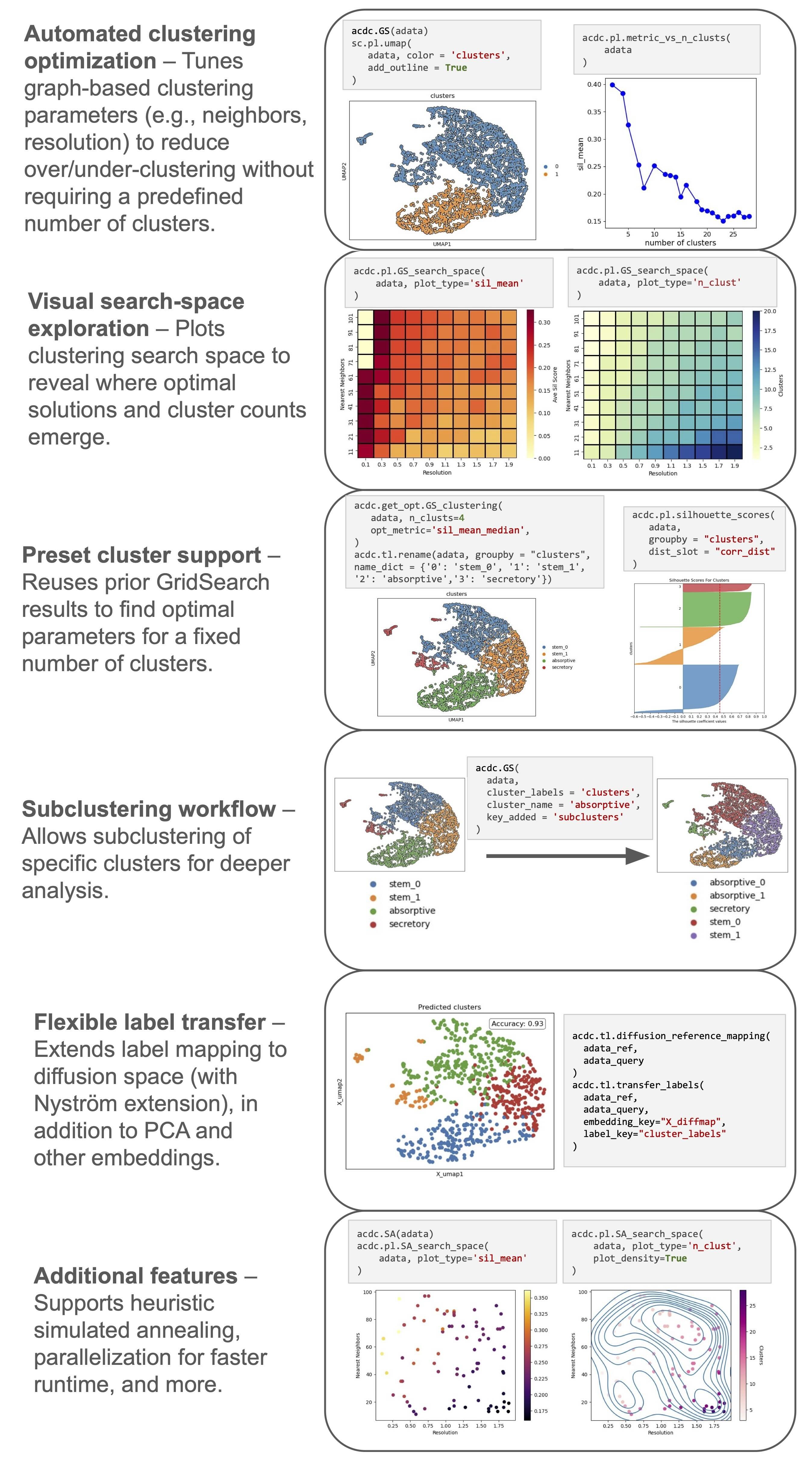
• Easy computation of optimal clustering solutions via unbiased parameter tuning
• Scanpy-compatible framework
• Built for speed, memory efficiency, and scalability with large scRNAseq datasets
• Straightforward subclustering for working with broad and distinct lineages simultaneously
Conceptual details of this procedure are available in the Methods section of Malagola, Vasciaveo, et al. Cell 2024
Tutorial 1: Clustering human single-cell RNA seq data with ACDC
Tutorial 2: Diffusion Map Embedding & Label Transfer with acdc_py
pip install acdc-pygit clone https://github.com/VasciaveoLab/acdc_py/
cd acdc_py
pip install -e .... Start playing around! 🎸
If you use acdc_py in your work, please cite the acdc_py publication as follows:
Protocol for automated graph-based clustering of single-cell RNA-seq data with application in mouse intestinal stem cells.
Wang, A. L., Zanella, L., Ochiai, Y., Golinelli, L., Califano, A., Malagola, E., & Vasciaveo, A.
STAR protocols 2025 Sep 19. doi: 10.1016/j.xpro.2025.104000.
- Malagola, E., Vasciaveo, A., Ochiai, Y., Kim, W., Zheng, B., Zanella, L., et al. (2024). Isthmus progenitor cells contribute to homeostatic cellular turnover and support regeneration following intestinal injury. Cell, 187(12), 3056–3071.
- Traag, V. A., Waltman, L., & Van Eck, N. J. (2019). From Louvain to Leiden: guaranteeing well-connected communities. Scientific Reports, 9(1), 5233.
- Wolf, F. A., Angerer, P., & Theis, F. J. (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biology, 19(1), 15.
- Xiang, Y., & Gong, X. G. (2000). Efficiency of generalized simulated annealing. Physical Review E, 62(3), 4473.
- Weiler, P., Lange, M., Klein, M., Pe’er, D., & Theis, F. (2024). CellRank 2: unified fate mapping in multiview single-cell data. Nature Methods, 21(7), 1196–1205.
- Alvarez, M. J., Shen, Y., Giorgi, F. M., Lachmann, A., Ding, B. B., Ye, B. H., et al. (2016). Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nature Genetics, 48(8), 838–847.
- Wang, A. L., Lin, Z., Zanella, L., Vlahos, L., Anglada Girotto, M., Zafar, A., et al. (2024). pyVIPER: A fast and scalable Python package for rank-based enrichment analysis of single-cell RNA-seq data. bioRxiv.
- Bradbury, J., Frostig, R., Hawkins, P., Johnson, M. J., Leary, C., Maclaurin, D., et al. (2018). JAX: Composable transformations of Python+NumPy programs (Version 0.3.13). GitHub repository. https://github.com/google/jax
- Lachmann, A., Giorgi, F. M., Lopez, G., & Califano, A. (2016). ARACNe-AP: gene network reverse engineering through adaptive partitioning inference of mutual information. Bioinformatics, 32(14), 2233–2235.
- Obradovic, A., Chowdhury, N., Haake, S. M., Ager, C., Wang, V., Vlahos, L., et al. (2021). Single-cell protein activity analysis identifies recurrence-associated renal tumor macrophages. Cell, 184(11), 2988–3005.
- Caliński, T., & Harabasz, J. (1974). A dendrite method for cluster analysis. Communications in Statistics – Theory and Methods, 3(1), 1–27.
- Davies, D. L., & Bouldin, D. W. (2009). A cluster separation measure. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2(2), 224–227.
- Cura, T. (2012). A particle swarm optimization approach to clustering. Expert Systems with Applications, 39(1), 1582–1588.
- Zhang, C., Ouyang, D., & Ning, J. (2010). An artificial bee colony approach for clustering. Expert Systems with Applications, 37(7), 4761–4767.
- Shelokar, P. S., Jayaraman, V. K., & Kulkarni, B. D. (2004). An ant colony approach for clustering. Analytica Chimica Acta, 509(2), 187–195.
This work was supported by grants from an NCI Outstanding Investigator Award (R35 CA197745) and the NIH Shared Instrumentation grants S10OD012351, S10OD021764, and S10OD032433, all to A.C. A.V. is supported by an Early Career Development Pilot Award from NIH/NCI Cancer Center, funded through the Cancer Center Support grant, P30CA013696.
Alessandro Vasciaveo - [email protected]
Alexander Wang - [email protected]
Luca Zanella - [email protected]