+  +
+
News! (2022) -- cleanlab made accessible for everybody, not just ML researchers (click to learn more)
--
-
-
- Nov 2022 📖 cleanlab 2.2.0 released! Added better algorithms for: label issues in multi-label classification, data with some classes absent, and estimating the number of label errors in a dataset. -
- Sep 2022 📖 cleanlab 2.1.0 released! Added support for: data labeled by multiple annotators in cleanlab.multiannotator, token classification with text data in cleanlab.token_classification, out-of-distribution detection in cleanlab.outlier, and CleanLearning with non-numpy-array data (e.g. pandas dataframes, tensorflow/pytorch datasets, etc) in cleanlab.classification.CleanLearning. -
- April 2022 📖 cleanlab 2.0.0 released! Lays foundations for this library to grow into a general-purpose data-centric AI toolkit. -
- March 2022 📖 Documentation migrated to new website: docs.cleanlab.ai with quickstart tutorials for image/text/audio/tabular data. -
- Feb 2022 💻 APIs simplified to make cleanlab accessible for everybody, not just ML researchers -
- Long-time cleanlab user? Here's how to migrate to cleanlab versions >= 2.0.0. -
News! (2021) -- cleanlab finds pervasive label errors in the most common ML datasets (click to learn more)
--
-
-
- Dec 2021 🎉 NeurIPS published the label errors paper (Northcutt, Athalye, & Mueller, 2021). -
- Apr 2021 🎉 Journal of AI Research published the confident learning paper (Northcutt, Jiang, & Chuang, 2021). -
- Mar 2021 😲 cleanlab used to find and fix label issues in 10 of the most common ML benchmark datasets, published in: NeurIPS 2021. Along with the paper (Northcutt, Athalye, & Mueller, 2021), the authors launched labelerrors.com where you can view the label issues in these datasets. -
News! (2020) -- cleanlab supports all OS, achieves state-of-the-art performance (click to learn more)
--
-
-
- Dec 2020 🎉 cleanlab supports NeurIPS workshop paper (Northcutt, Athalye, & Lin, 2020). -
- Dec 2020 🤖 cleanlab supports PU learning. -
- Feb 2020 🤖 cleanlab now natively supports Mac, Linux, and Windows. -
- Feb 2020 🤖 cleanlab now supports Co-Teaching (Han et al., 2018). -
- Jan 2020 🎉 cleanlab achieves state-of-the-art on CIFAR-10 with noisy labels. Code to reproduce: examples/cifar10. This is a great place to see how to use cleanlab on real datasets (with predicted probabilities from trained model already precomputed for you). -
+  +
+
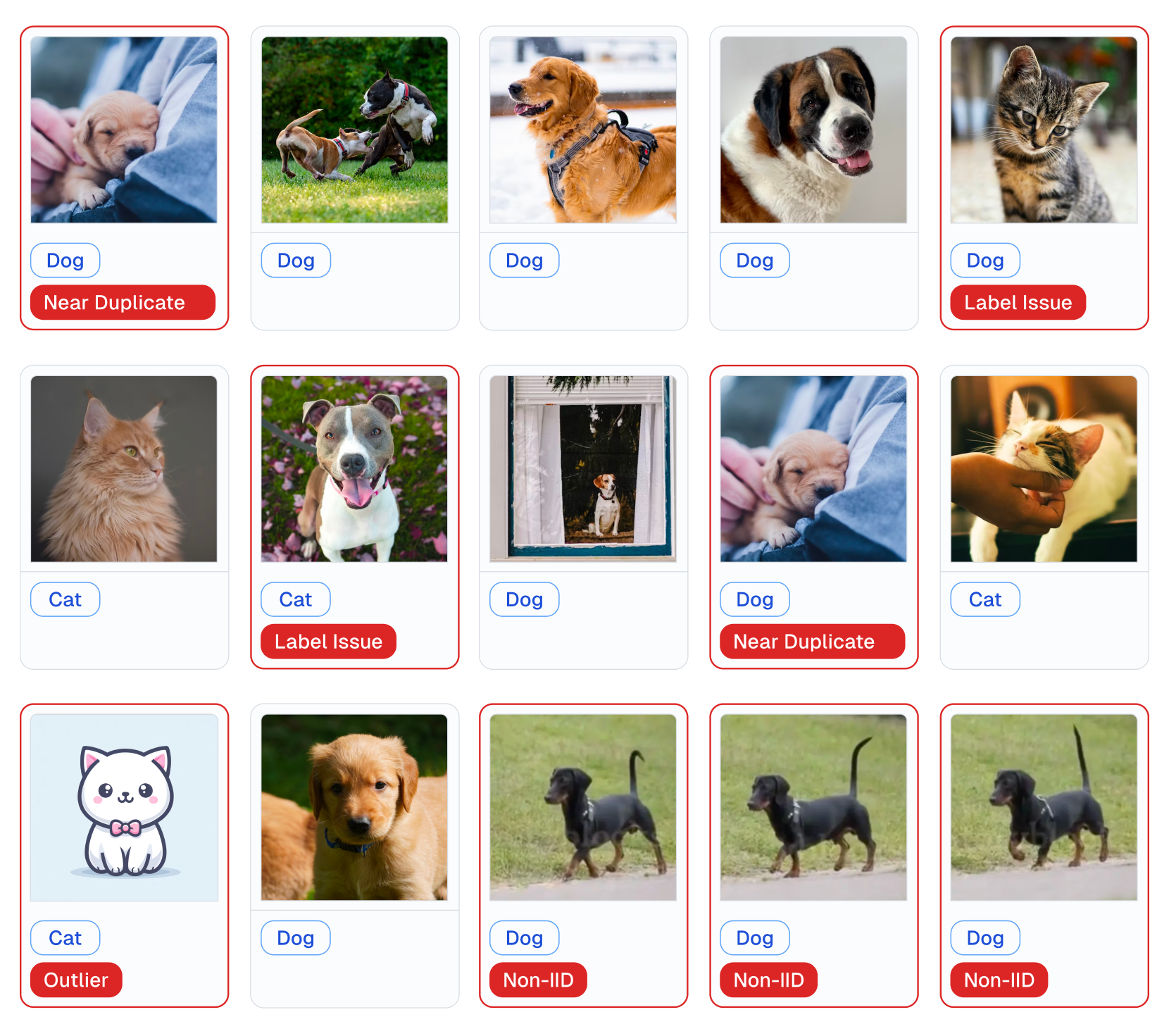
+ Examples of various issues in Cat/Dog dataset automatically detected by cleanlab (with 1 line of code).
- @@ -103,11 +71,23 @@ cleanlab supports Linux, macOS, and Windows and runs on Python 3.7+. - Get started [here](https://docs.cleanlab.ai/)! Install via `pip` or `conda` as described [here](https://docs.cleanlab.ai/). - Developers who install the bleeding-edge from source should refer to [this master branch documentation](https://docs.cleanlab.ai/master/index.html). +- For help, check out our detailed [FAQ](https://docs.cleanlab.ai/stable/tutorials/faq.html), [Github Issues](https://github.com/cleanlab/cleanlab/issues?q=is%3Aissue), or [Slack](https://cleanlab.ai/slack). We welcome any questions! + +**Practicing data-centric AI can look like this:** +1. Train initial ML model on original dataset. +2. Utilize this model to diagnose data issues (via cleanlab methods) and improve the dataset. +3. Train the same model on the improved dataset. +4. Try various modeling techniques to further improve performance. + +Most folks jump from Step 1 → 4, but you may achieve big gains without *any* change to your modeling code by using cleanlab! +Continuously boost performance by iterating Steps 2 → 4 (and try to evaluate with *cleaned* data). + +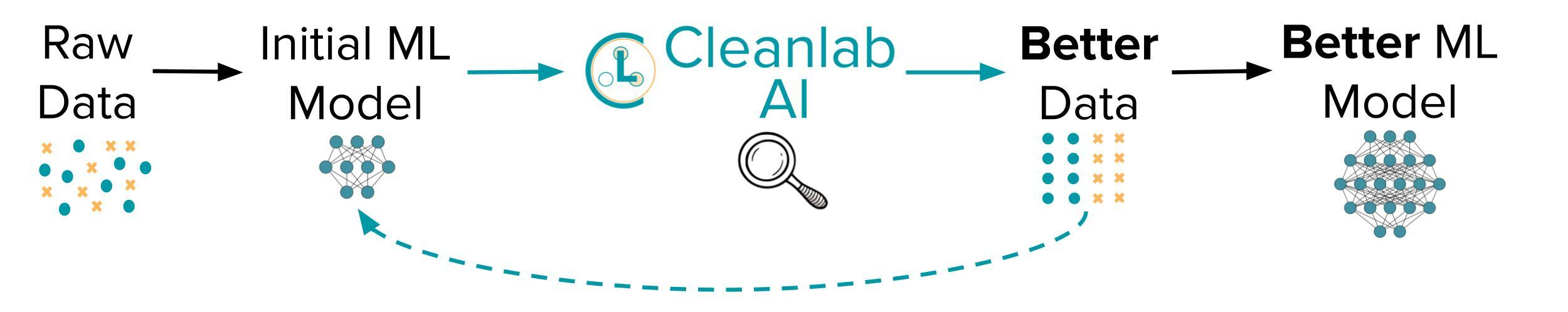 ## Use cleanlab with any model for most ML tasks -All features of cleanlab work with **any dataset** and **any model**. Yes, any model: scikit-learn, PyTorch, Tensorflow, Keras, JAX, HuggingFace, MXNet, XGBoost, etc. +All features of cleanlab work with **any dataset** and **any model**. Yes, any model: PyTorch, Tensorflow, Keras, JAX, HuggingFace, OpenAI, XGBoost, scikit-learn, etc. If you use a sklearn-compatible classifier, all cleanlab methods work out-of-the-box.
@@ -117,7 +97,7 @@ It’s also easy to use your favorite non-sklearn-compatible model (click to
cleanlab can find label issues from any model's predicted class probabilities if you can produce them yourself.
-Some other cleanlab functionality requires your model to be sklearn-compatible.
+Some cleanlab functionality may require your model to be sklearn-compatible.
There's nothing you need to do if your model already has `.fit()`, `.predict()`, and `.predict_proba()` methods.
Otherwise, just wrap your custom model into a Python class that inherits the `sklearn.base.BaseEstimator`:
@@ -150,300 +130,23 @@ cl.predict(test_data)
More details are provided in documentation of [cleanlab.classification.CleanLearning](https://docs.cleanlab.ai/stable/cleanlab/classification.html).
-Note, some libraries exist to give you sklearn-compatibility for free. For PyTorch, check out the [skorch](https://skorch.readthedocs.io/) Python library which will wrap your PyTorch model into a sklearn-compatible model ([example](https://docs.cleanlab.ai/stable/tutorials/image.html)). For TensorFlow/Keras, check out [SciKeras](https://www.adriangb.com/scikeras/) ([example](https://docs.cleanlab.ai/stable/tutorials/text.html)) or [our own Keras wrapper](https://docs.cleanlab.ai/stable/cleanlab/experimental/keras.html). Many libraries also already offer a special scikit-learn API, for example: [XGBoost](https://xgboost.readthedocs.io/en/stable/python/python_api.html#module-xgboost.sklearn) or [LightGBM](https://lightgbm.readthedocs.io/en/latest/pythonapi/lightgbm.LGBMClassifier.html).
+Note, some libraries exist to give you sklearn-compatibility for free. For PyTorch, check out the [skorch](https://skorch.readthedocs.io/) Python library which will wrap your PyTorch model into a sklearn-compatible model ([example](https://docs.cleanlab.ai/stable/tutorials/image.html)). For TensorFlow/Keras, check out our [Keras wrapper](https://docs.cleanlab.ai/stable/cleanlab/models/keras.html). Many libraries also already offer a special scikit-learn API, for example: [XGBoost](https://xgboost.readthedocs.io/en/stable/python/python_api.html#module-xgboost.sklearn) or [LightGBM](https://lightgbm.readthedocs.io/en/latest/pythonapi/lightgbm.LGBMClassifier.html).
-Reproducing results in Confident Learning paper -(click to learn more) -
-- -For additional details, check out the: [confidentlearning-reproduce repository](https://github.com/cgnorthcutt/confidentlearning-reproduce). - -### State of the Art Learning with Noisy Labels in CIFAR - -A step-by-step guide to reproduce these results is available [here](https://github.com/cleanlab/examples/tree/master/contrib/v1/cifar10). This guide is also a good tutorial for using cleanlab on any large dataset. You'll need to `git clone` -[confidentlearning-reproduce](https://github.com/cgnorthcutt/confidentlearning-reproduce) which contains the data and files needed to reproduce the CIFAR-10 results. - -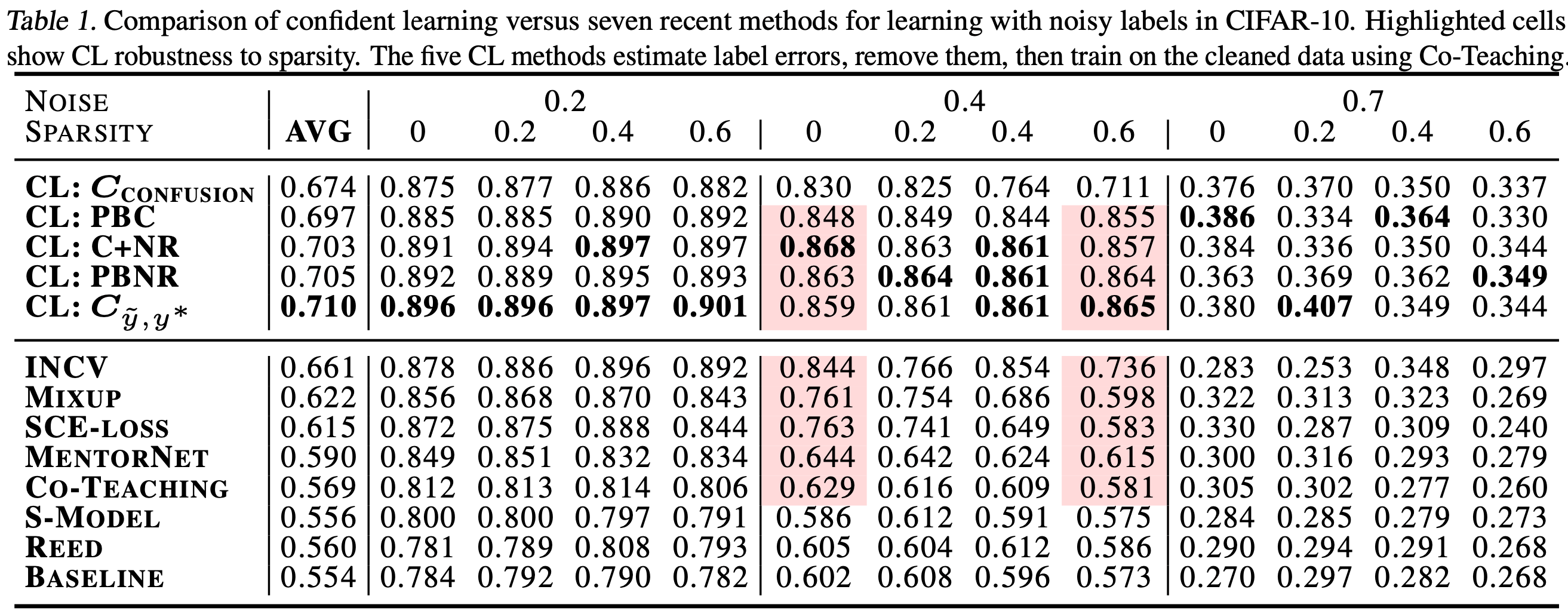 - -Comparison of confident learning (CL), as implemented in cleanlab, versus seven recent methods for learning with noisy labels in CIFAR-10. Highlighted cells show CL robustness to sparsity. The five CL methods estimate label issues, remove them, then train on the cleaned data using [Co-Teaching](https://github.com/cleanlab/cleanlab/blob/master/cleanlab/experimental/coteaching.py). - -Observe how cleanlab (i.e. the CL method) is robust to large sparsity in label noise whereas prior art tends to reduce in performance for increased sparsity, as shown by the red highlighted regions. This is important because real-world label noise is often sparse, e.g. a tiger is likely to be mislabeled as a lion, but not as most other classes like airplane, bathtub, and microwave. - -### Find label issues in ImageNet - -Use cleanlab to identify \~100,000 label errors in the 2012 ILSVRC ImageNet training dataset: [examples/imagenet](https://github.com/cleanlab/examples/tree/master/contrib/v1/imagenet). - -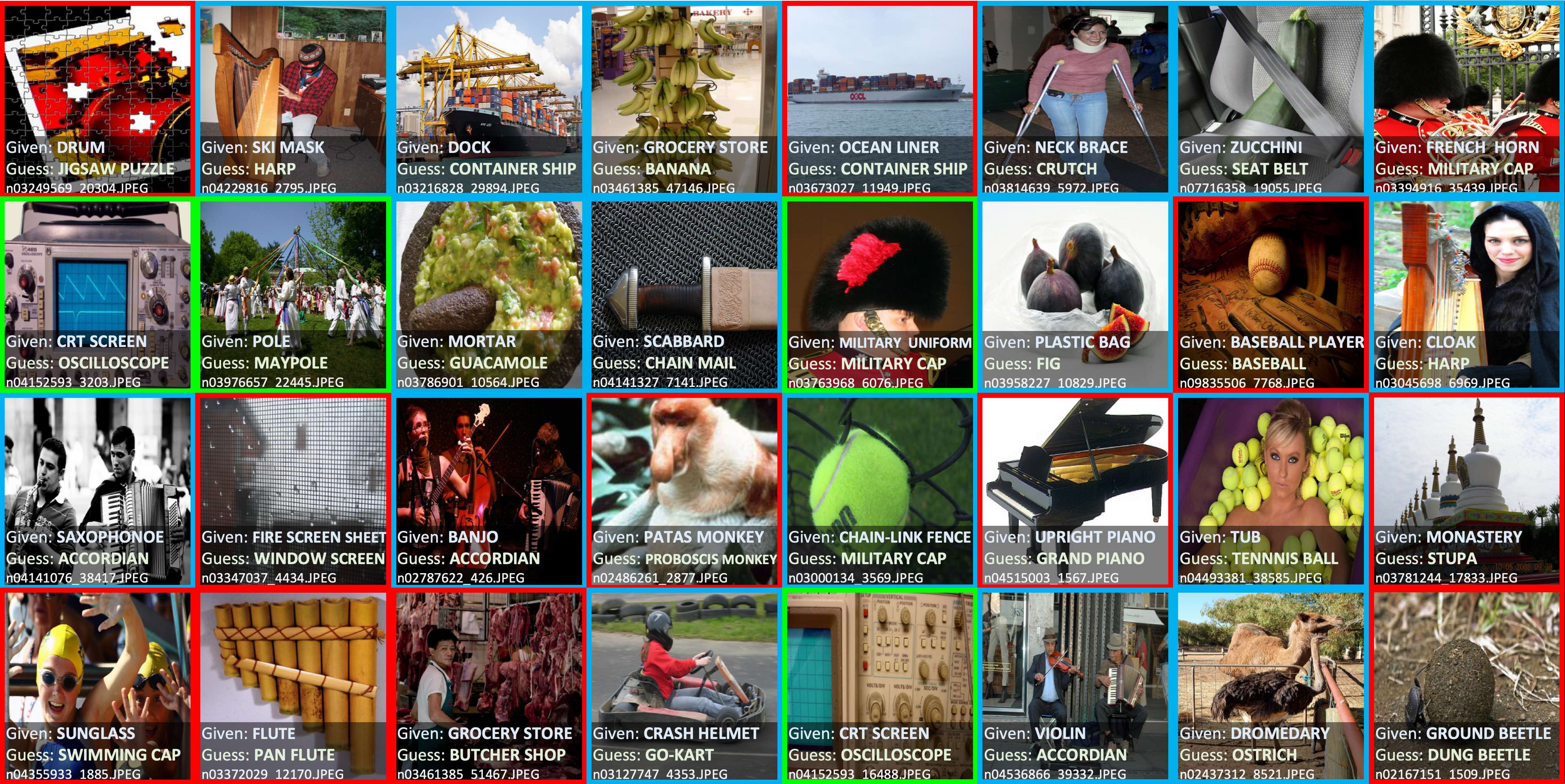 - -Label issues in ImageNet train set found via cleanlab. Label Errors are boxed in red. Ontological issues in green. Multi-label images in blue. - -### Find Label Errors in MNIST - -Use cleanlab to identify \~50 label errors in the MNIST dataset: [examples/mnist](https://github.com/cleanlab/examples/tree/master/contrib/v1/mnist). - -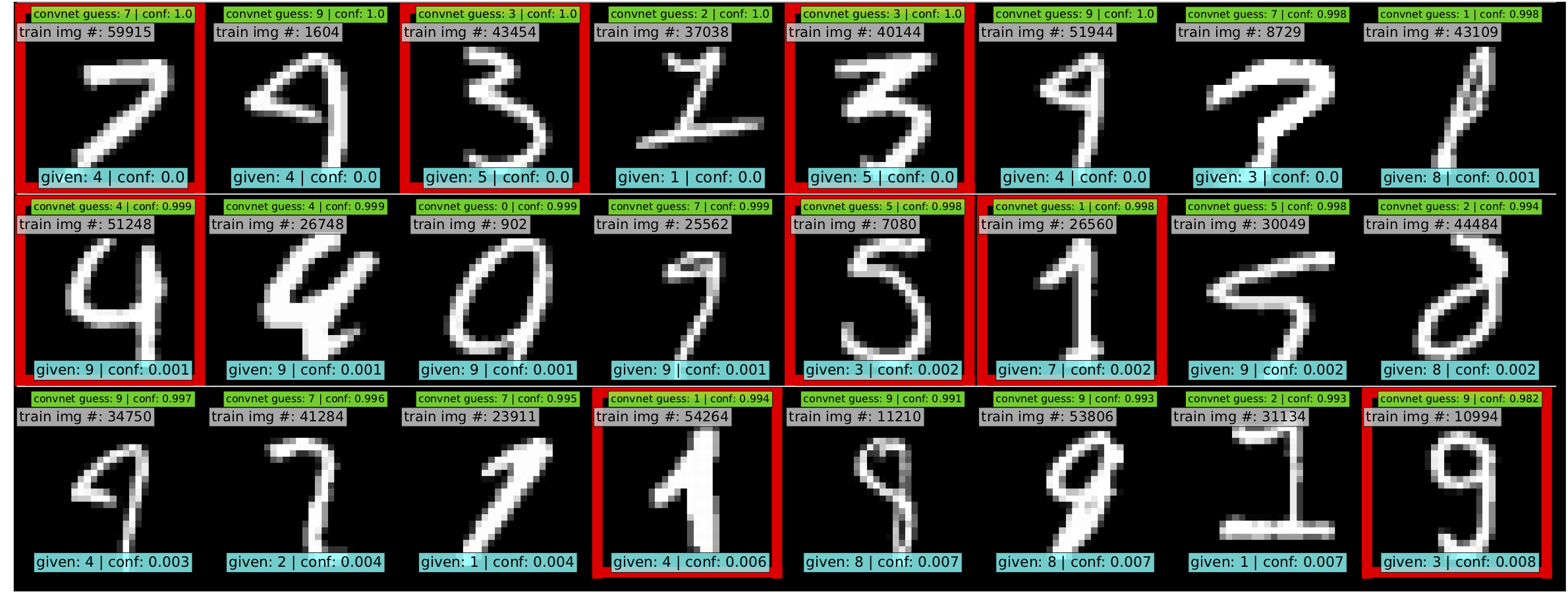 - -Top 24 least-confident labels in the original MNIST **train** dataset, algorithmically identified via cleanlab. Examples are ordered left-right, top-down by increasing self-confidence (predicted probability that the **given** label is correct), denoted **conf** in teal. The most-likely correct label (with largest predicted probability) is in green. Overt label errors highlighted in red. - -
-
-Learning with noisy labels across 4 data distributions and 9 classifiers -(click to learn more) -
-- -cleanlab is a general tool that can learn with noisy labels regardless of dataset distribution or classifier type: [examples/classifier\_comparison](https://github.com/cleanlab/examples/blob/master/classifier_comparison/classifier_comparison.ipynb). - -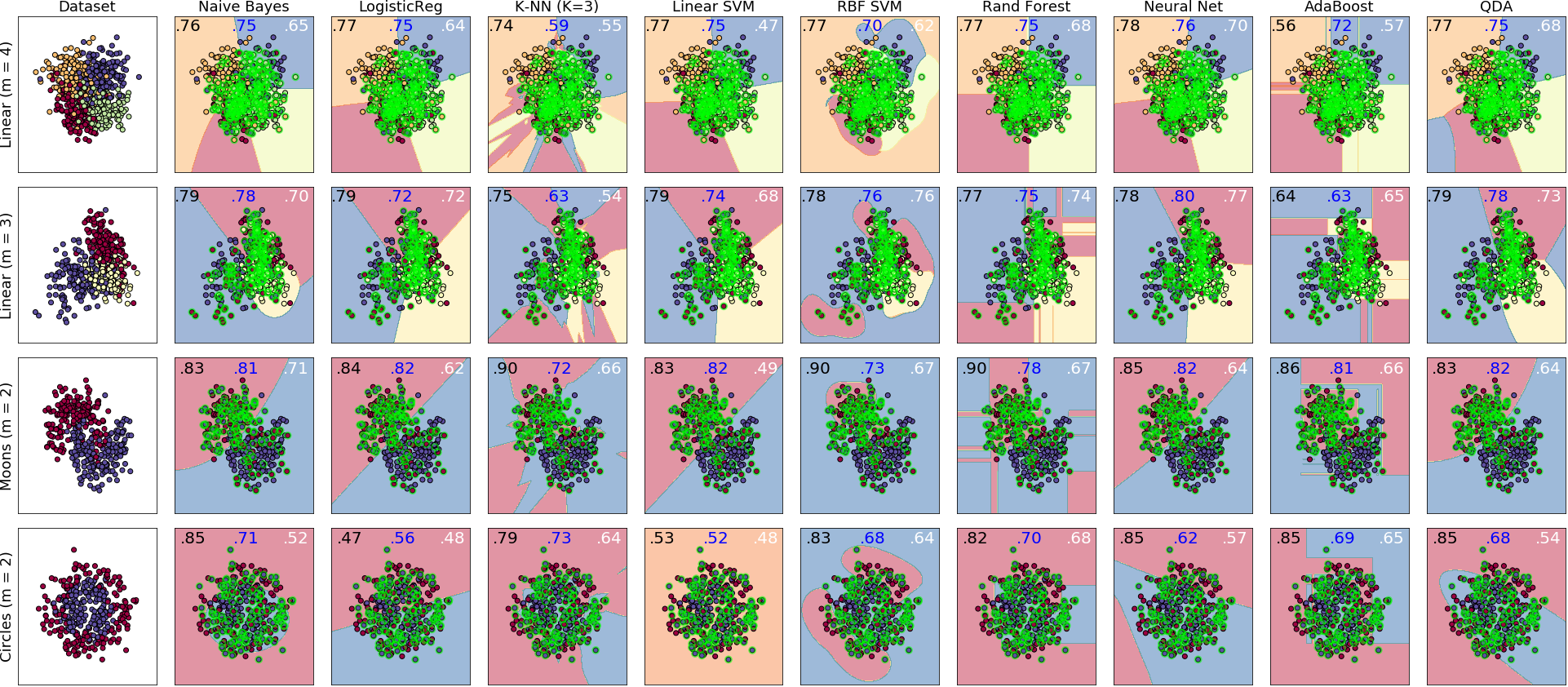 - -Each sub-figure above depicts the decision boundary learned using [cleanlab.classification.CleanLearning](https://github.com/cleanlab/cleanlab/blob/master/cleanlab/classification.py#L141) in the presence of extreme (\~35%) label errors (circled in green). Label noise is class-conditional (not uniformly random). Columns are organized by the classifier used, except the left-most column which depicts the ground-truth data distribution. Rows are organized by dataset. - -Each sub-figure depicts accuracy scores on a test set (with correct non-noisy labels) as decimal values: - -* LEFT (in black): The classifier test accuracy trained with perfect labels (no label errors). -* MIDDLE (in blue): The classifier test accuracy trained with noisy labels using cleanlab. -* RIGHT (in white): The baseline classifier test accuracy trained with noisy labels. - -As an example, the table below is the noise matrix (noisy channel) *P(s | y) -characterizing the label noise for the first dataset row in the figure. *s* represents the observed noisy labels and *y* represents the latent, true labels. The trace of this matrix is 2.6. A trace of 4 implies no label noise. A cell in this matrix is read like: "Around 38% of true underlying '3' labels were randomly flipped to '2' labels in the -observed dataset." - -| `p(label︱y)` | y=0 | y=1 | y=2 | y=3 | -|--------------|------|------|------|------| -| label=0 | 0.55 | 0.01 | 0.07 | 0.06 | -| label=1 | 0.22 | 0.87 | 0.24 | 0.02 | -| label=2 | 0.12 | 0.04 | 0.64 | 0.38 | -| label=3 | 0.11 | 0.08 | 0.05 | 0.54 | - -
-
-ML research using cleanlab -(click to learn more) -
-- -Researchers may find some components of this package useful for evaluating algorithms for ML with noisy labels. For additional details/notation, refer to [the Confident Learning paper](https://jair.org/index.php/jair/article/view/12125). - -### Methods to Standardize Research with Noisy Labels - -cleanlab supports a number of functions to generate noise for benchmarking and standardization in research. This next example shows how to generate valid, class-conditional, uniformly random noisy channel matrices: - -``` python -# Generate a valid (necessary conditions for learnability are met) noise matrix for any trace > 1 -from cleanlab.benchmarking.noise_generation import generate_noise_matrix_from_trace -noise_matrix=generate_noise_matrix_from_trace( - K=number_of_classes, - trace=float_value_greater_than_1_and_leq_K, - py=prior_of_y_actual_labels_which_is_just_an_array_of_length_K, - frac_zero_noise_rates=float_from_0_to_1_controlling_sparsity, -) - -# Check if a noise matrix is valid (necessary conditions for learnability are met) -from cleanlab.benchmarking.noise_generation import noise_matrix_is_valid -is_valid=noise_matrix_is_valid( - noise_matrix, - prior_of_y_which_is_just_an_array_of_length_K, -) -``` - -For a given noise matrix, this example shows how to generate noisy labels. Methods can be seeded for reproducibility. - -``` python -# Generate noisy labels using the noise_marix. Guarantees exact amount of noise in labels. -from cleanlab.benchmarking.noise_generation import generate_noisy_labels -s_noisy_labels = generate_noisy_labels(y_hidden_actual_labels, noise_matrix) - -# This package is a full of other useful methods for learning with noisy labels. -# The tutorial stops here, but you don't have to. Inspect method docstrings for full docs. -``` - -
-
-cleanlab for advanced users -(click to learn more) -
-- -Many methods and their default parameters are not covered here. Check out the [documentation for the master branch version](https://docs.cleanlab.ai/master/) for the full suite of features supported by the cleanlab API. - -## Use any custom model's predicted probabilities to find label errors in 1 line of code - -pred_probs (num_examples x num_classes matrix of predicted probabilities) should already be computed on your own, with any classifier. For best results, pred_probs should be obtained in a holdout/out-of-sample manner (e.g. via cross-validation). -* cleanlab can do this for you via [`cleanlab.count.estimate_cv_predicted_probabilities`](https://docs.cleanlab.ai/master/cleanlab/count.html)] -* Tutorial with more info: [[here](https://docs.cleanlab.ai/stable/tutorials/pred_probs_cross_val.html)] -* Examples how to compute pred_probs with: [[CNN image classifier (PyTorch)](https://docs.cleanlab.ai/stable/tutorials/image.html)], [[NN text classifier (TensorFlow)](https://docs.cleanlab.ai/stable/tutorials/text.html)] - -```python -# label issues are ordered by likelihood of being an error. First index is most likely error. -from cleanlab.filter import find_label_issues - -ordered_label_issues = find_label_issues( # One line of code! - labels=numpy_array_of_noisy_labels, - pred_probs=numpy_array_of_predicted_probabilities, - return_indices_ranked_by='normalized_margin', # Orders label issues - ) -``` - -Pre-computed **out-of-sample** predicted probabilities for CIFAR-10 train set are available: [here](https://github.com/cleanlab/examples/tree/master/contrib/v1/cifar10#pre-computed-psx-for-every-noise--sparsity-condition). - -## Fully characterize label noise and uncertainty in your dataset. - -*s* denotes a random variable that represents the observed, noisy label and *y* denotes a random variable representing the hidden, actual labels. Both *s* and *y* take any of the m classes as values. The cleanlab package supports different levels of granularity for computation depending on the needs of the user. Because of this, we support multiple alternatives, all no more than a few lines, to estimate these latent distribution arrays, enabling the user to reduce computation time by only computing what they need to compute, as seen in the examples below. - -Throughout these examples, you’ll see a variable called *confident\_joint*. The confident joint is an m x m matrix (m is the number of classes) that counts, for every observed, noisy class, the number of examples that confidently belong to every latent, hidden class. It counts the number of examples that we are confident are labeled correctly or incorrectly for every pair of observed and unobserved classes. The confident joint is an unnormalized estimate of the complete-information latent joint distribution, *Ps,y*. - -The label flipping rates are denoted *P(s | y)*, the inverse rates are *P(y | s)*, and the latent prior of the unobserved, true labels, *p(y)*. - -Most of the methods in the **cleanlab** package start by first estimating the *confident\_joint*. You can learn more about this in the [confident learning paper](https://arxiv.org/abs/1911.00068). - -### Option 1: Compute the confident joint and predicted probs first. Stop if that’s all you need. - -``` python -from cleanlab.count import estimate_latent -from cleanlab.count import estimate_confident_joint_and_cv_pred_proba - -# Compute the confident joint and the n x m predicted probabilities matrix (pred_probs), -# for n examples, m classes. Stop here if all you need is the confident joint. -confident_joint, pred_probs = estimate_confident_joint_and_cv_pred_proba( - X=X_train, - labels=train_labels_with_errors, - clf=logreg(), # default, you can use any classifier -) - -# Estimate latent distributions: p(y) as est_py, P(s|y) as est_nm, and P(y|s) as est_inv -est_py, est_nm, est_inv = estimate_latent( - confident_joint, - labels=train_labels_with_errors, -) -``` - -### Option 2: Estimate the latent distribution matrices in a single line of code. - -``` python -from cleanlab.count import estimate_py_noise_matrices_and_cv_pred_proba -est_py, est_nm, est_inv, confident_joint, pred_probs = estimate_py_noise_matrices_and_cv_pred_proba( - X=X_train, - labels=train_labels_with_errors, -) -``` - -### Option 3: Skip computing the predicted probabilities if you already have them. - -``` python -# Already have pred_probs? (n x m matrix of predicted probabilities) -# For example, you might get them from a pre-trained model (like resnet on ImageNet) -# With the cleanlab package, you estimate directly with pred_probs. -from cleanlab.count import estimate_py_and_noise_matrices_from_probabilities -est_py, est_nm, est_inv, confident_joint = estimate_py_and_noise_matrices_from_probabilities( - labels=train_labels_with_errors, - pred_probs=pred_probs, -) -``` - -## Completely characterize label noise in a dataset: - -The joint probability distribution of noisy and true labels, *P(s,y)*, completely characterizes label noise with a class-conditional *m x m* matrix. - -``` python -from cleanlab.count import estimate_joint -joint = estimate_joint( - labels=noisy_labels, - pred_probs=probabilities, - confident_joint=None, # Provide if you have it already -) -``` - -
-
-Positive-Unlabeled Learning -(click to learn more) -
-- -Positive-Unlabeled (PU) learning (in which your data only contains a few positively labeled examples with the rest unlabeled) is just a special case of [CleanLearning](https://github.com/cleanlab/cleanlab/blob/master/cleanlab/classification.py#L141) when one of the classes has no error. `P` stands for the positive class and **is assumed to have zero label errors** and `U` stands for unlabeled data, but in practice, we just assume the `U` class is a noisy negative class that actually contains some positive examples. Thus, the goal of PU learning is to (1) estimate the proportion of negatively labeled examples that actually belong to the positive class (see`fraction\_noise\_in\_unlabeled\_class` in the last example), (2) find the errors (see last example), and (3) train on clean data (see first example below). cleanlab does all three, taking into account that there are no label errors in whichever class you specify as positive. - -There are two ways to use cleanlab for PU learning. We'll look at each here. - -Method 1. If you are using the cleanlab classifier [CleanLearning()](https://github.com/cleanlab/cleanlab/blob/master/cleanlab/classification.py#L141), and your dataset has exactly two classes (positive = 1, and negative = 0), PU -learning is supported directly in cleanlab. You can perform PU learning like this: - -``` python -from cleanlab.classification import CleanLearning -from sklearn.linear_model import LogisticRegression -# Wrap around any classifier. Yup, you can use sklearn/pyTorch/TensorFlow/FastText/etc. -pu_class = 0 # Should be 0 or 1. Label of class with NO ERRORS. (e.g., P class in PU) -cl = CleanLearning(clf=LogisticRegression(), pulearning=pu_class) -cl.fit(X=X_train_data, labels=train_noisy_labels) -# Estimate the predictions you would have gotten by training with *no* label errors. -predicted_test_labels = cl.predict(X_test) -``` - -Method 2. However, you might be using a more complicated classifier that doesn't work well with [CleanLearning](https://github.com/cleanlab/cleanlab/blob/master/cleanlab/classification.py#L141) (see this example for CIFAR-10). Or you might have 3 or more classes. Here's how to use cleanlab for PU learning in this situation. To let cleanlab know which class has no error (in standard PU learning, this is the P class), you need to set the threshold for that class to 1 (1 means the probability that the labels of that class are correct is 1, i.e. that class has no -error). Here's the code: - -``` python -import numpy as np -# K is the number of classes in your dataset -# pred_probs are the cross-validated predicted probabilities. -# s is the array/list/iterable of noisy labels -# pu_class is a 0-based integer for the class that has no label errors. -thresholds = np.asarray([np.mean(pred_probs[:, k][s == k]) for k in range(K)]) -thresholds[pu_class] = 1.0 -``` - -Now you can use cleanlab however you were before. Just be sure to pass in `thresholds` as a parameter wherever it applies. For example: - -``` python -# Uncertainty quantification (characterize the label noise -# by estimating the joint distribution of noisy and true labels) -cj = compute_confident_joint(s, pred_probs, thresholds=thresholds, ) -# Now the noise (cj) has been estimated taking into account that some class(es) have no error. -# We can use cj to find label errors like this: -indices_of_label_issues = find_label_issues(s, pred_probs, confident_joint=cj, ) - -# In addition to label issues, cleanlab can find the fraction of noise in the unlabeled class. -# First we need the inv_noise_matrix which contains P(y|s) (proportion of mislabeling). -_, _, inv_noise_matrix = estimate_latent(confident_joint=cj, labels=s, ) -# Because inv_noise_matrix contains P(y|s), p (y = anything | labels = pu_class) should be 0 -# because the prob(true label is something else | example is in pu_class) is 0. -# What's more interesting is p(y = anything | s is not put_class), or in the binary case -# this translates to p(y = pu_class | s = 1 - pu_class) because pu_class is 0 or 1. -# So, to find the fraction_noise_in_unlabeled_class, for binary, you just compute: -fraction_noise_in_unlabeled_class = inv_noise_matrix[pu_class][1 - pu_class] -``` - -Now that you have `indices_of_label_errors`, you can remove those label issues and train on clean data (or only remove some of the label issues and iteratively use confident learning / cleanlab to improve results). - -
-
CROWDLAB for data with multiple annotators (NeurIPS '22) (click to show bibtex)
@inproceedings{goh2022crowdlab, - title={Utilizing supervised models to infer consensus labels and their quality from data with multiple annotators}, + title={CROWDLAB: Supervised learning to infer consensus labels and quality scores for data with multiple annotators}, author={Goh, Hui Wen and Tkachenko, Ulyana and Mueller, Jonas}, booktitle={NeurIPS Human in the Loop Learning Workshop}, year={2022} @@ -521,21 +224,75 @@ cleanlab is based on peer-reviewed research. Here are relevant papers to cite ifActiveLab: Active learning with data re-labeling (ICLR '23) (click to show bibtex)
+ + @inproceedings{goh2023activelab, + title={ActiveLab: Active Learning with Re-Labeling by Multiple Annotators}, + author={Goh, Hui Wen and Mueller, Jonas}, + booktitle={ICLR Workshop on Trustworthy ML}, + year={2023} + } + +Incorrect Annotations in Multi-Label Classification (ICLR '23) (click to show bibtex)
+ + @inproceedings{thyagarajan2023multilabel, + title={Identifying Incorrect Annotations in Multi-Label Classification Data}, + author={Thyagarajan, Aditya and Snorrason, Elías and Northcutt, Curtis and Mueller, Jonas}, + booktitle={ICLR Workshop on Trustworthy ML}, + year={2023} + } + +Detecting Dataset Drift and Non-IID Sampling (ICML '23) (click to show bibtex)
+ + @inproceedings{cummings2023drift, + title={Detecting Dataset Drift and Non-IID Sampling via k-Nearest Neighbors}, + author={Cummings, Jesse and Snorrason, Elías and Mueller, Jonas}, + booktitle={ICML Workshop on Data-centric Machine Learning Research}, + year={2023} + } + +Detecting Errors in Numerical Data (ICML '23) (click to show bibtex)
+ + @inproceedings{zhou2023errors, + title={Detecting Errors in Numerical Data via any Regression Model}, + author={Zhou, Hang and Mueller, Jonas and Kumar, Mayank and Wang, Jane-Ling and Lei, Jing}, + booktitle={ICML Workshop on Data-centric Machine Learning Research}, + year={2023} + } + +
+  +
+