-
Notifications
You must be signed in to change notification settings - Fork 12
/
Copy path07_high_dimensional_data.Rmd
1734 lines (1194 loc) · 45.6 KB
/
07_high_dimensional_data.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
502
503
504
505
506
507
508
509
510
511
512
513
514
515
516
517
518
519
520
521
522
523
524
525
526
527
528
529
530
531
532
533
534
535
536
537
538
539
540
541
542
543
544
545
546
547
548
549
550
551
552
553
554
555
556
557
558
559
560
561
562
563
564
565
566
567
568
569
570
571
572
573
574
575
576
577
578
579
580
581
582
583
584
585
586
587
588
589
590
591
592
593
594
595
596
597
598
599
600
601
602
603
604
605
606
607
608
609
610
611
612
613
614
615
616
617
618
619
620
621
622
623
624
625
626
627
628
629
630
631
632
633
634
635
636
637
638
639
640
641
642
643
644
645
646
647
648
649
650
651
652
653
654
655
656
657
658
659
660
661
662
663
664
665
666
667
668
669
670
671
672
673
674
675
676
677
678
679
680
681
682
683
684
685
686
687
688
689
690
691
692
693
694
695
696
697
698
699
700
701
702
703
704
705
706
707
708
709
710
711
712
713
714
715
716
717
718
719
720
721
722
723
724
725
726
727
728
729
730
731
732
733
734
735
736
737
738
739
740
741
742
743
744
745
746
747
748
749
750
751
752
753
754
755
756
757
758
759
760
761
762
763
764
765
766
767
768
769
770
771
772
773
774
775
776
777
778
779
780
781
782
783
784
785
786
787
788
789
790
791
792
793
794
795
796
797
798
799
800
801
802
803
804
805
806
807
808
809
810
811
812
813
814
815
816
817
818
819
820
821
822
823
824
825
826
827
828
829
830
831
832
833
834
835
836
837
838
839
840
841
842
843
844
845
846
847
848
849
850
851
852
853
854
855
856
857
858
859
860
861
862
863
864
865
866
867
868
869
870
871
872
873
874
875
876
877
878
879
880
881
882
883
884
885
886
887
888
889
890
891
892
893
894
895
896
897
898
899
900
901
902
903
904
905
906
907
908
909
910
911
912
913
914
915
916
917
918
919
920
921
922
923
924
925
926
927
928
929
930
931
932
933
934
935
936
937
938
939
940
941
942
943
944
945
946
947
948
949
950
951
952
953
954
955
956
957
958
959
960
961
962
963
964
965
966
967
968
969
970
971
972
973
974
975
976
977
978
979
980
981
982
983
984
985
986
987
988
989
990
991
992
993
994
995
996
997
998
999
1000
# High-dimensional data {#machine_learning}
```{r include = FALSE}
# Caching this markdown file
#knitr::opts_chunk$set(cache = TRUE)
```
## The Big Picture
- The rise of high-dimensional data. The new data frontiers in social sciences---text ([Gentzkow et al. 2019](https://web.stanford.edu/~gentzkow/research/text-as-data.pdf); [Grimmer and Stewart 2013](https://www.jstor.org/stable/pdf/24572662.pdf?casa_token=SQdSI4R_VdwAAAAA:4QiVLhCXqr9f0qNMM9U75EL5JbDxxnXxUxyIfDf0U8ZzQx9szc0xVqaU6DXG4nHyZiNkvcwGlgD6H0Lxj3y0ULHwgkf1MZt8-9TPVtkEH9I4AHgbTg)) and and image ([Joo and Steinert-Threlkeld 2018](https://arxiv.org/pdf/1810.01544))---are all high-dimensional data.
- 1000 common English words for 30-word tweets: $1000^{30}$ similar to N of atoms in the universe ([Gentzkow et al. 2019](https://web.stanford.edu/~gentzkow/research/text-as-data.pdf))
- Belloni, Alexandre, Victor Chernozhukov, and Christian Hansen. ["High-dimensional methods and inference on structural and treatment effects."](https://pubs.aeaweb.org/doi/pdfplus/10.1257/jep.28.2.29) *Journal of Economic Perspectives 28*, no. 2 (2014): 29-50.
- The rise of the new approach: statistics + computer science = machine learning
- Statistical inference
- $y$ <- some probability models (e.g., linear regression, logistic regression) <- $x$
- $y$ = $X\beta$ + $\epsilon$
- The goal is to estimate $\beta$
- Machine learning
- $y$ <- unknown <- $x$
- $y$ <-> decision trees, neutral nets <-> $x$
- For the main idea behind prediction modeling, see Breiman, Leo (Berkeley stat faculty who passed away in 2005). ["Statistical modeling: The two cultures (with comments and a rejoinder by the author)."](https://projecteuclid.org/euclid.ss/1009213726) *Statistical science* 16, no. 3 (2001): 199-231.
- "The problem is to find an algorithm $f(x)$ such that for future $x$ in a test set, $f(x)$ will be a good predictor of $y$."
- "There are **two cultures** in the use of statistical modeling to reach conclusions from data. One assumes that the data are generated by a **given** **stochastic data model**. The other uses **algorithmic models** and treats the data mechanism as **unknown**."
- How ML differs from econometrics?
- A review by Athey, Susan, and Guido W. Imbens. ["Machine learning methods that economists should know about."](https://www.annualreviews.org/doi/full/10.1146/annurev-economics-080217-053433) *Annual Review of Economics* 11 (2019): 685-725.
- Stat:
- Specifying a target (i.e., an estimand)
- Fitting a model to data using an objective function (e.g., the sum of squared errors)
- Reporting point estimates (effect size) and standard errors (uncertainty)
- Validation by yes-no using goodness-of-fit tests and residual examination
- ML:
- Developing algorithms (estimating *f(x)*)
- Prediction power, not structural/causal parameters
- Basically, high-dimensional data statistics (N < P)
- The major problem is to avoid ["the curse of dimensionality"](https://en.wikipedia.org/wiki/Curse_of_dimensionality) ([too many features - > overfitting](https://towardsdatascience.com/the-curse-of-dimensionality-50dc6e49aa1e))
- Validation: out-of-sample comparisons (cross-validation) not in-sample goodness-of-fit measures
- So, it's curve-fitting, but the primary focus is unseen (test data), not seen data (training data)
- A quick review on ML lingos for those trained in econometrics
- Sample to estimate parameters = Training sample
- Estimating the model = Being trained
- Regressors, covariates, or predictors = Features
- Regression parameters = weights
- Prediction problems = Supervised (some $y$ are known) + Unsupervised ($y$ unknown)
. Many images in this chapter come from vas3k blog.](https://i.vas3k.ru/7w9.jpg)
](https://i.vas3k.ru/7vz.jpg)
](https://i.vas3k.ru/7vx.jpg)
](https://i.vas3k.ru/7w1.jpg)
## Dataset
- [Heart disease data from UCI](https://archive.ics.uci.edu/ml/datasets/heart+Disease)
- One of the popular datasets used in machine learning competitions
```{r}
# Load packages
## CRAN packages
pacman::p_load(
here,
tidyverse,
tidymodels,
doParallel, # parallel processing
patchwork, # arranging ggplots
remotes,
SuperLearner,
vip,
tidymodels,
glmnet,
xgboost,
rpart,
ranger,
conflicted
)
remotes::install_github("ck37/ck37r")
conflicted::conflict_prefer("filter", "dplyr")
```
```{r}
## Jae's custom functions
source(here("functions", "ml_utils.r"))
# Import the dataset
data_original <- read_csv(here("data", "heart.csv"))
glimpse(data_original)
# Createa a copy
data <- data_original
theme_set(theme_minimal())
```
## Workflow
```{r echo=FALSE, results='asis'}
pacman::p_load(glue)
workflow_list <- c(
"Preprocessing",
"Model building",
"Model fitting",
"Model evaluation",
"Model tuning",
"Prediction"
)
glue("- {seq(workflow_list)}. {workflow_list}")
```
## tidymodels
- Like `tidyverse`, `tidymodels` is a collection of packages.
- [`rsample`](https://rsample.tidymodels.org/): for data splitting
- [`recipes`](https://recipes.tidymodels.org/index.html): for pre-processing
- [`parsnip`](https://www.tidyverse.org/blog/2018/11/parsnip-0-0-1/): for model building
- [`tune`](https://github.com/tidymodels/tune): hyperparameter tuning
- [`yardstick`](https://github.com/tidymodels/yardstick): for model evaluations
- [`workflows`](https://github.com/tidymodels/workflows): for bundling a pieplne that bundles together preprocessing, modeling, and post-processing requests
- Why taking a tidyverse approach to machine learning?
- Benefits
- Readable code
- Reusable data structures
- Extendable code

> tidymodels are an **integrated, modular, extensible** set of packages that implement a framework that facilitates creating predicative stochastic models. - Joseph Rickert@RStudio
- Currently, 238 models are [available](https://topepo.github.io/caret/available-models.html)
- The following materials are based on [the machine learning with tidymodels workshop](https://github.com/dlab-berkeley/Machine-Learning-with-tidymodels) I developed for D-Lab. [The original workshop](https://github.com/dlab-berkeley/Machine-Learning-in-R) was designed by [Chris Kennedy](https://ck37.com/) and [Evan Muzzall](https://dlab.berkeley.edu/people/evan-muzzall.
## Pre-processing
- [`recipes`](https://recipes.tidymodels.org/index.html): for pre-processing
- [`textrecipes`](https://github.com/tidymodels/textrecipes) for text pre-processing
- Step 1: `recipe()` defines target and predictor variables (ingredients).
- Step 2: `step_*()` defines preprocessing steps to be taken (recipe).
The preprocessing steps list draws on the vignette of the [`parsnip`](https://www.tidymodels.org/find/parsnip/) package.
- dummy: Also called one-hot encoding
- zero variance: Removing columns (or features) with a single unique value
- impute: Imputing missing values
- decorrelate: Mitigating correlated predictors (e.g., principal component analysis)
- normalize: Centering and/or scaling predictors (e.g., log scaling). Scaling matters because many algorithms (e.g., lasso) are scale-variant (except tree-based algorithms). Remind you that normalization (sensitive to outliers) = $\frac{X - X_{min}}{X_{max} - X_{min}}$ and standardization (not sensitive to outliers) = $\frac{X - \mu}{\sigma}$
- transform: Making predictors symmetric
- Step 3: `prep()` prepares a dataset to base each step on.
- Step 4: `bake()` applies the preprocessing steps to your datasets.
In this course, we focus on two preprocessing tasks.
- One-hot encoding (creating dummy/indicator variables)
```{r}
# Turn selected numeric variables into factor variables
data <- data %>%
dplyr::mutate(across(c("sex", "ca", "cp", "slope", "thal"), as.factor))
glimpse(data)
```
- Imputation
```{r}
# Check missing values
map_df(data, ~ is.na(.) %>% sum())
# Add missing values
data$oldpeak[sample(seq(data), size = 10)] <- NA
# Check missing values
# Check the number of missing values
data %>%
map_df(~ is.na(.) %>% sum())
# Check the rate of missing values
data %>%
map_df(~ is.na(.) %>% mean())
```
### Regression setup
#### Outcome variable
```{r}
# Continuous variable
data$age %>% class()
```
#### Data splitting using random sampling
```{r}
# for reproducibility
set.seed(1234)
# split
split_reg <- initial_split(data, prop = 0.7)
# training set
raw_train_x_reg <- training(split_reg)
# test set
raw_test_x_reg <- testing(split_reg)
```
#### recipe
```{r}
# Regression recipe
rec_reg <- raw_train_x_reg %>%
# Define the outcome variable
recipe(age ~ .) %>%
# Median impute oldpeak column
step_impute_median(oldpeak) %>%
# Expand "sex", "ca", "cp", "slope", and "thal" features out into dummy variables (indicators).
step_dummy(c("sex", "ca", "cp", "slope", "thal"))
# Prepare a dataset to base each step on
prep_reg <- rec_reg %>% prep(retain = TRUE)
```
```{r}
# x features
train_x_reg <- juice(prep_reg, all_predictors())
test_x_reg <- bake(
object = prep_reg,
new_data = raw_test_x_reg, all_predictors()
)
# y variables
train_y_reg <- juice(prep_reg, all_outcomes())$age %>% as.numeric()
test_y_reg <- bake(prep_reg, raw_test_x_reg, all_outcomes())$age %>% as.numeric()
# Checks
names(train_x_reg) # Make sure there's no age variable!
class(train_y_reg) # Make sure this is a continuous variable!
```
- Note that other imputation methods are also available.
```{r}
grep("impute", ls("package:recipes"), value = TRUE)
```
- You can also create your own `step_` functions. For more information, see [tidymodels.org](https://www.tidymodels.org/learn/develop/recipes/).
### Classification setup
#### Outcome variable
```{r}
data$target %>% class()
data$target <- as.factor(data$target)
data$target %>% class()
```
#### Data splitting using stratified random sampling
```{r}
# split
split_class <- initial_split(data %>%
mutate(target = as.factor(target)),
prop = 0.7,
strata = target
)
# training set
raw_train_x_class <- training(split_class)
# testing set
raw_test_x_class <- testing(split_class)
```
#### recipe
```{r}
# Classification recipe
rec_class <- raw_train_x_class %>%
# Define the outcome variable
recipe(target ~ .) %>%
# Median impute oldpeak column
step_impute_median(oldpeak) %>%
# Expand "sex", "ca", "cp", "slope", and "thal" features out into dummy variables (indicators).
step_normalize(age) %>%
step_dummy(c("sex", "ca", "cp", "slope", "thal"))
# Prepare a dataset to base each step on
prep_class <- rec_class %>% prep(retain = TRUE)
```
```{r}
# x features
train_x_class <- juice(prep_class, all_predictors())
test_x_class <- bake(prep_class, raw_test_x_class, all_predictors())
# y variables
train_y_class <- juice(prep_class, all_outcomes())$target %>% as.factor()
test_y_class <- bake(prep_class, raw_test_x_class, all_outcomes())$target %>% as.factor()
# Checks
names(train_x_class) # Make sure there's no target variable!
class(train_y_class) # Make sure this is a factor variable!
```
## Supervised learning
x -> f - > y (defined)
### OLS and Lasso
#### parsnip
- Build models (`parsnip`)
1. Specify a model
2. Specify an engine
3. Specify a mode
```{r}
# OLS spec
ols_spec <- linear_reg() %>% # Specify a model
set_engine("lm") %>% # Specify an engine: lm, glmnet, stan, keras, spark
set_mode("regression") # Declare a mode: regression or classification
```
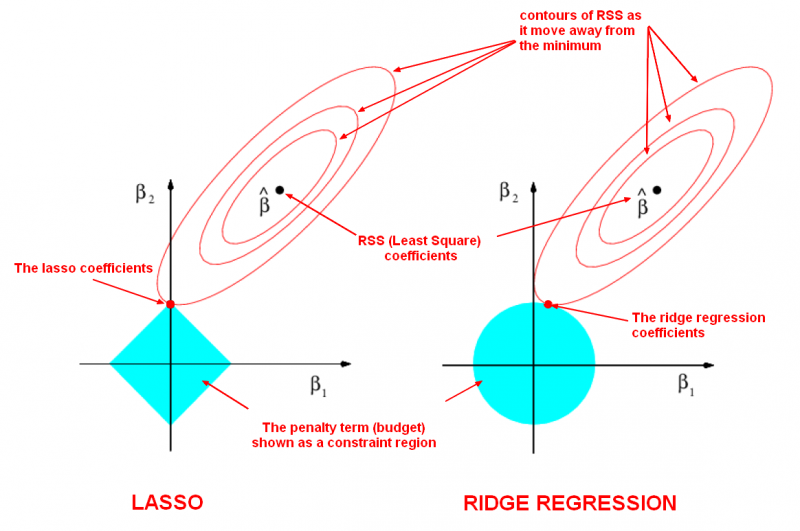
Lasso is one of the regularization techniques along with ridge and elastic-net.
```{r}
# Lasso spec
lasso_spec <- linear_reg(
penalty = 0.1, # tuning hyperparameter
mixture = 1
) %>% # 1 = lasso, 0 = ridge
set_engine("glmnet") %>%
set_mode("regression")
# If you don't understand parsnip arguments
lasso_spec %>% translate() # See the documentation
```
- Fit models
```{r}
ols_fit <- ols_spec %>%
fit_xy(x = train_x_reg, y = train_y_reg)
# fit(train_y_reg ~ ., train_x_reg) # When you data are not preprocessed
lasso_fit <- lasso_spec %>%
fit_xy(x = train_x_reg, y = train_y_reg)
```
#### yardstick
- Visualize model fits
```{r}
map2(list(ols_fit, lasso_fit), c("OLS", "Lasso"), visualize_fit)
```
```{r}
# Define performance metrics
metrics <- yardstick::metric_set(rmse, mae, rsq)
# Evaluate many models
evals <- purrr::map(list(ols_fit, lasso_fit), evaluate_reg) %>%
reduce(bind_rows) %>%
mutate(type = rep(c("OLS", "Lasso"), each = 3))
# Visualize the test results
evals %>%
ggplot(aes(x = fct_reorder(type, .estimate), y = .estimate)) +
geom_point() +
labs(
x = "Model",
y = "Estimate"
) +
facet_wrap(~ glue("{toupper(.metric)}"), scales = "free_y")
```
- For more information, read [Tidy Modeling with R](https://www.tmwr.org/) by Max Kuhn and Julia Silge.
#### tune
**Hyper**parameters are parameters that control the learning process.
##### tune ingredients
- Search space for hyperparameters
1. Grid search: a grid of hyperparameters
2. Random search: random sample points from a bounded domain

```{r}
# tune() = placeholder
tune_spec <- linear_reg(
penalty = tune(), # tuning hyperparameter
mixture = 1
) %>% # 1 = lasso, 0 = ridge
set_engine("glmnet") %>%
set_mode("regression")
tune_spec
# penalty() searches 50 possible combinations
lambda_grid <- grid_regular(penalty(), levels = 50)
```
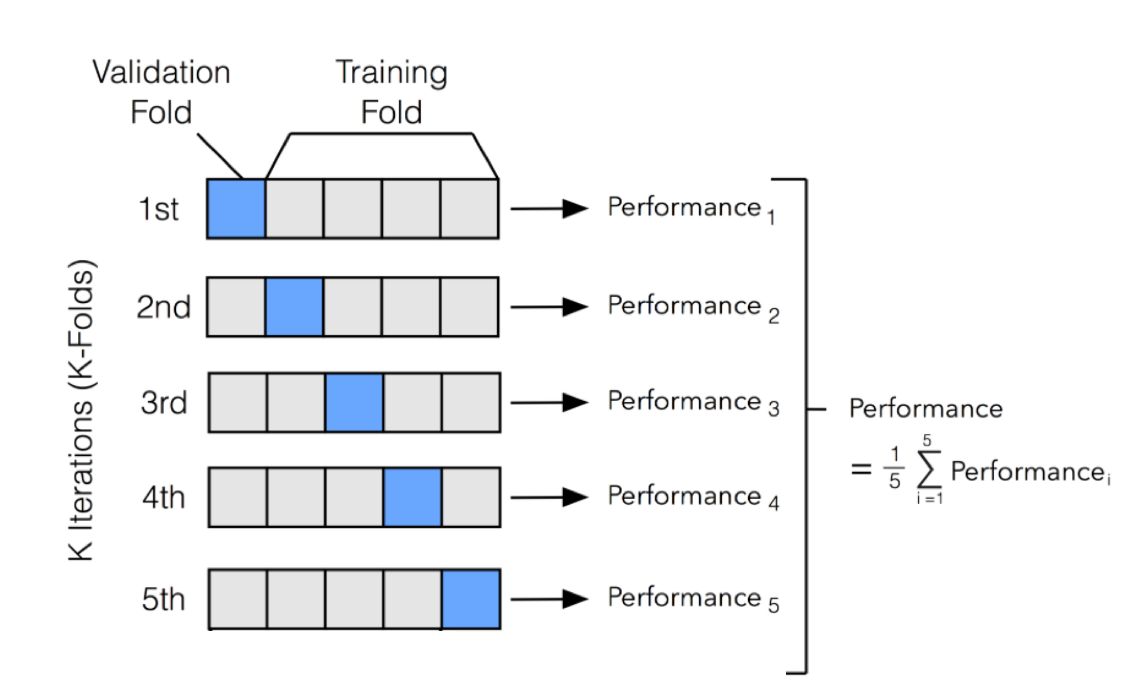
```{r}
# 10-fold cross-validation
set.seed(1234) # for reproducibility
rec_folds <- vfold_cv(train_x_reg %>% bind_cols(tibble(age = train_y_reg)))
```
##### Add these elements to a workflow
```{r}
# Workflow
rec_wf <- workflow() %>%
add_model(tune_spec) %>%
add_formula(age ~ .)
```
```{r}
# Tuning results
rec_res <- rec_wf %>%
tune_grid(
resamples = rec_folds,
grid = lambda_grid
)
```
##### Visualize
```{r}
# Visualize
rec_res %>%
collect_metrics() %>%
ggplot(aes(penalty, mean, col = .metric)) +
geom_errorbar(aes(
ymin = mean - std_err,
ymax = mean + std_err
),
alpha = 0.3
) +
geom_line(size = 2) +
scale_x_log10() +
labs(x = "log(lambda)") +
facet_wrap(~ glue("{toupper(.metric)}"),
scales = "free",
nrow = 2
) +
theme(legend.position = "none")
```
##### Select
```{r}
conflict_prefer("filter", "dplyr")
top_rmse <- show_best(rec_res, metric = "rmse")
best_rmse <- select_best(rec_res, metric = "rmse")
best_rmse
glue('The RMSE of the intiail model is
{evals %>%
filter(type == "Lasso", .metric == "rmse") %>%
select(.estimate) %>%
round(2)}')
glue('The RMSE of the tuned model is {rec_res %>%
collect_metrics() %>%
filter(.metric == "rmse") %>%
arrange(mean) %>%
dplyr::slice(1) %>%
select(mean) %>%
round(2)}')
```
- Finalize your workflow and visualize [variable importance](https://koalaverse.github.io/vip/articles/vip.html)
```{r}
finalize_lasso <- rec_wf %>%
finalize_workflow(best_rmse)
finalize_lasso %>%
fit(train_x_reg %>% bind_cols(tibble(age = train_y_reg))) %>%
pull_workflow_fit() %>%
vip::vip()
```
##### Test fit
- Apply the tuned model to the test dataset
```{r}
test_fit <- finalize_lasso %>%
fit(test_x_reg %>% bind_cols(tibble(age = test_y_reg)))
evaluate_reg(test_fit)
```
### Decision tree
#### parsnip
- Build a model
1. Specify a model
2. Specify an engine
3. Specify a mode
```{r}
# workflow
tree_wf <- workflow() %>% add_formula(target ~ .)
# spec
tree_spec <- decision_tree(
# Mode
mode = "classification",
# Tuning hyperparameters
cost_complexity = NULL,
tree_depth = NULL
) %>%
set_engine("rpart") # rpart, c5.0, spark
tree_wf <- tree_wf %>% add_model(tree_spec)
```
- Fit a model
```{r}
tree_fit <- tree_wf %>% fit(train_x_class %>% bind_cols(tibble(target = train_y_class)))
```
#### yardstick
- Let's formally test prediction performance.
1. Confusion matrix
A confusion matrix is often used to describe the performance of a classification model. The below example is based on a binary classification model.
| | Predicted: YES | Predicted: NO |
|-----------------|---------------------|---------------------|
| **Actual: YES** | True positive (TP) | False negative (FN) |
| **Actual: NO** | False positive (FP) | True negative (TN) |
2. Metrics
- `accuracy`: The proportion of the data predicted correctly ($\frac{TP + TN}{total}$). 1 - accuracy = misclassification rate.
- `precision`: Positive predictive value. *When the model predicts yes, how correct is it?* ($\frac{TP}{TP + FP}$)
- `recall` (sensitivity): True positive rate (e.g., healthy people healthy). *When the actual value is yes, how often does the model predict yes?* ($\frac{TP}{TP + FN}$)
- `F-score`: A weighted average between precision and recall.
- `ROC Curve` (receiver operating characteristic curve): a plot that shows the relationship between true and false positive rates at different classification thresholds. y-axis indicates the true positive rate and x-axis indicates the false positive rate. What matters is the AUC (Area under the ROC Curve), which is a cumulative probability function of ranking a random "positive" - "negative" pair (for the probability of AUC, see [this blog post](https://www.alexejgossmann.com/auc/)).

- To learn more about other metrics, check out the yardstick package [references](https://yardstick.tidymodels.org/reference/index.html).
```{r}
# Define performance metrics
metrics <- yardstick::metric_set(accuracy, precision, recall)
# Visualize
tree_fit_viz_metr <- visualize_class_eval(tree_fit)
tree_fit_viz_metr
tree_fit_viz_mat <- visualize_class_conf(tree_fit)
tree_fit_viz_mat
```
#### tune
##### tune ingredients
Decisions trees tend to overfit. We need to consider two things to reduce this problem: how to split and when to stop a tree.
- **complexity parameter**: a high CP means a simple decision tree with few splits.
- **tree_depth**
```{r}
tune_spec <- decision_tree(
cost_complexity = tune(), # how to split
tree_depth = tune(), # when to stop
mode = "classification"
) %>%
set_engine("rpart")
tree_grid <- grid_regular(cost_complexity(),
tree_depth(),
levels = 5
) # 2 hyperparameters -> 5*5 = 25 combinations
tree_grid %>%
count(tree_depth)
# 10-fold cross-validation
set.seed(1234) # for reproducibility
tree_folds <- vfold_cv(train_x_class %>% bind_cols(tibble(target = train_y_class)),
strata = target
)
```
##### Add these elements to a workflow
```{r}
# Update workflow
tree_wf <- tree_wf %>% update_model(tune_spec)
# Determine the number of cores
no_cores <- detectCores() - 1
# Initiate
cl <- makeCluster(no_cores)
registerDoParallel(cl)
# Tuning results
tree_res <- tree_wf %>%
tune_grid(
resamples = tree_folds,
grid = tree_grid,
metrics = metrics
)
```
##### Visualize
- The following plot draws on the [vignette](https://www.tidymodels.org/start/tuning/) of the tidymodels package.
```{r}
tree_res %>%
collect_metrics() %>%
mutate(tree_depth = factor(tree_depth)) %>%
ggplot(aes(cost_complexity, mean, col = .metric)) +
geom_point(size = 3) +
# Subplots
facet_wrap(~tree_depth,
scales = "free",
nrow = 2
) +
# Log scale x
scale_x_log10(labels = scales::label_number()) +
# Discrete color scale
scale_color_viridis_d(option = "plasma", begin = .9, end = 0) +
labs(
x = "Cost complexity",
col = "Tree depth",
y = NULL
) +
coord_flip()
```
##### Select
```{r}
# Optimal hyperparameter
best_tree <- select_best(tree_res, "recall")
# Add the hyperparameter to the workflow
finalize_tree <- tree_wf %>%
finalize_workflow(best_tree)
```
```{r}
tree_fit_tuned <- finalize_tree %>%
fit(train_x_class %>% bind_cols(tibble(target = train_y_class)))
# Metrics
(tree_fit_viz_metr + labs(title = "Non-tuned")) / (visualize_class_eval(tree_fit_tuned) + labs(title = "Tuned"))
# Confusion matrix
(tree_fit_viz_mat + labs(title = "Non-tuned")) / (visualize_class_conf(tree_fit_tuned) + labs(title = "Tuned"))
```
- Visualize variable importance
```{r}
tree_fit_tuned %>%
pull_workflow_fit() %>%
vip::vip()
```
##### Test fit
- Apply the tuned model to the test dataset
```{r}
test_fit <- finalize_tree %>%
fit(test_x_class %>% bind_cols(tibble(target = test_y_class)))
evaluate_class(test_fit)
```
In the next subsection, we will learn variants of ensemble models that improve decision tree models by putting models together.
### Bagging (Random forest)
Key idea applied across all ensemble models (bagging, boosting, and stacking):
single learner -> N learners (N > 1)
Many learners could perform better than a single learner as this approach reduces the **variance** of a single estimate and provides more stability.
Here we focus on the difference between bagging and boosting. In short, boosting may reduce bias while increasing variance. On the other hand, bagging may reduce variance but has nothing to do with bias. Please check out [What is the difference between Bagging and Boosting?](https://quantdare.com/what-is-the-difference-between-bagging-and-boosting/) by aporras.
**bagging**
- Data: Training data will be randomly sampled with replacement (bootstrapping samples + drawing random **subsets** of features for training individual trees)
- Learning: Building models in parallel (independently)
- Prediction: Simple average of the estimated responses (majority vote system)
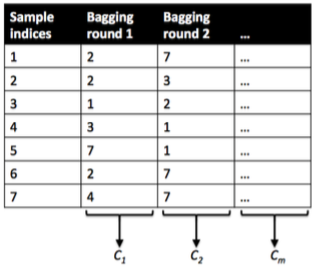
**boosting**
- Data: Weighted training data will be random sampled
- Learning: Building models sequentially (mispredicted cases would receive more weights)
- Prediction: Weighted average of the estimated responses
#### parsnip
- Build a model
1. Specify a model
2. Specify an engine
3. Specify a mode
```{r}
# workflow
rand_wf <- workflow() %>% add_formula(target ~ .)
# spec
rand_spec <- rand_forest(
# Mode
mode = "classification",
# Tuning hyperparameters
mtry = NULL, # The number of predictors to available for splitting at each node
min_n = NULL, # The minimum number of data points needed to keep splitting nodes
trees = 500
) %>% # The number of trees
set_engine("ranger",
# We want the importance of predictors to be assessed.
seed = 1234,
importance = "permutation"
)
rand_wf <- rand_wf %>% add_model(rand_spec)
```
- Fit a model
```{r}
rand_fit <- rand_wf %>% fit(train_x_class %>% bind_cols(tibble(target = train_y_class)))
```
#### yardstick
```{r}
# Define performance metrics
metrics <- yardstick::metric_set(accuracy, precision, recall)
rand_fit_viz_metr <- visualize_class_eval(rand_fit)
rand_fit_viz_metr
```
- Visualize the confusion matrix.
```{r}
rand_fit_viz_mat <- visualize_class_conf(rand_fit)
rand_fit_viz_mat
```
#### tune
##### tune ingredients
We focus on the following two hyperparameters:
- `mtry`: The number of predictors available for splitting at each node.
- `min_n`: The minimum number of data points needed to keep splitting nodes.
```{r}
tune_spec <-
rand_forest(
mode = "classification",
# Tuning hyperparameters
mtry = tune(),
min_n = tune()
) %>%
set_engine("ranger",
seed = 1234,
importance = "permutation"
)
rand_grid <- grid_regular(mtry(range = c(1, 10)),
min_n(range = c(2, 10)),
levels = 5
)
rand_grid %>%
count(min_n)
```
```{r}
# 10-fold cross-validation
set.seed(1234) # for reproducibility
rand_folds <- vfold_cv(train_x_class %>% bind_cols(tibble(target = train_y_class)),
strata = target
)
```
##### Add these elements to a workflow
```{r}
# Update workflow
rand_wf <- rand_wf %>% update_model(tune_spec)
# Tuning results
rand_res <- rand_wf %>%
tune_grid(
resamples = rand_folds,
grid = rand_grid,
metrics = metrics
)
```
##### Visualize
```{r}
rand_res %>%
collect_metrics() %>%
mutate(min_n = factor(min_n)) %>%
ggplot(aes(mtry, mean, color = min_n)) +
# Line + Point plot
geom_line(size = 1.5, alpha = 0.6) +
geom_point(size = 2) +
# Subplots
facet_wrap(~.metric,
scales = "free",
nrow = 2
) +
# Log scale x
scale_x_log10(labels = scales::label_number()) +
# Discrete color scale
scale_color_viridis_d(option = "plasma", begin = .9, end = 0) +
labs(
x = "The number of predictors to be sampled",
col = "The minimum number of data points needed for splitting",
y = NULL