diff --git a/.Rbuildignore b/.Rbuildignore
index b60a653..7ce4f93 100644
--- a/.Rbuildignore
+++ b/.Rbuildignore
@@ -13,3 +13,4 @@ docs/
^Makefile$
extradata/
revdep/
+^CRAN-SUBMISSION$
diff --git a/DESCRIPTION b/DESCRIPTION
index 804ec89..e296d87 100644
--- a/DESCRIPTION
+++ b/DESCRIPTION
@@ -1,7 +1,7 @@
Package: rsparse
Type: Package
Title: Statistical Learning on Sparse Matrices
-Version: 0.5.0
+Version: 0.5.1
Authors@R: c(
person("Dmitriy", "Selivanov", role=c("aut", "cre", "cph"), email="ds@rexy.ai",
comment = c(ORCID = "0000-0001-5413-1506")),
@@ -57,5 +57,5 @@ Suggests:
StagedInstall: TRUE
URL: https://github.com/rexyai/rsparse
BugReports: https://github.com/rexyai/rsparse/issues
-RoxygenNote: 7.1.1
+RoxygenNote: 7.2.1
NeedsCompilation: yes
diff --git a/NEWS.md b/NEWS.md
index c5950e3..e5cb2a5 100644
--- a/NEWS.md
+++ b/NEWS.md
@@ -1,21 +1,30 @@
-- 2021-10-17 - `v0.5.0`
- - reworked non-negative matrix factorization with brand-new Coordinate Descent solver for OLS
- - WRMF can model user, item and global biases
- - various performance improvements
-- 2020-04-01 - `v0.4.0`
- - updated docs with roxygen2 7.1
- - added `ScaleNormalize` transformer
- - added sparse*float S4 methods
-- 2019-07-17 - `v0.3.3.2`
- - faster `find_top_product()` - avoid BLAS and openmp thread contention
- - correctly identify openmp on OSX
- - fixed issue with CRAN 'rcnst' check
- - use `install_name_tool` hook in the `.onLoad()` - changes location of the `float.so` for CRAN binary installation - see #25
-- 2019-04-14 - `v0.3.3.1`
- - fixed out of bound memory access as reported by CRAN UBSAN
- - added ability to init GloVe embeddings with user provided values
-- 2019-03-16 - - `v0.3.3`
- - added methods to natively slice CSR matrices without converting them to triplet/CSC
-- 2018-10-25
- - add GloVe matrix factorization (adapted from `text2vec`)
- - link to `float` package - credits to @snoweye and @wrathematics
+# rsparse 0.5.1 (2022-09-11)
+- update `configure` script, thanks to @david-cortes, see #73
+- minor fixes in WRMF
+- update docs with new roxygen2 to pass CRAN checks
+- update NEWS.md ro follow CRAN format
+
+# rsparse 0.5.0 (2021-10-17)
+- reworked non-negative matrix factorization with brand-new Coordinate Descent solver for OLS
+- WRMF can model user, item and global biases
+- various performance improvements
+
+# rsparse 0.4.0 (2020-04-01)
+- updated docs with roxygen2 7.1
+- added `ScaleNormalize` transformer
+- added sparse*float S4 methods
+
+# rsparse 0.3.3.2 (2019-07-17)
+- faster `find_top_product()` - avoid BLAS and openmp thread contention
+- correctly identify openmp on OSX
+- fixed issue with CRAN 'rcnst' check
+- use `install_name_tool` hook in the `.onLoad()` - changes location of the `float.so` for CRAN binary installation - see #25
+
+# rsparse 0.3.3.1 (2019-04-14)
+- fixed out of bound memory access as reported by CRAN UBSAN
+- added ability to init GloVe embeddings with user provided values
+
+# rsparse 0.3.3 (2019-03-16)
+- added methods to natively slice CSR matrices without converting them to triplet/CSC
+- add GloVe matrix factorization (adapted from `text2vec`)
+- link to `float` package - credits to @snoweye and @wrathematics
diff --git a/R/model_FTRL.R b/R/model_FTRL.R
index 472acd2..d6ddf7d 100644
--- a/R/model_FTRL.R
+++ b/R/model_FTRL.R
@@ -147,7 +147,7 @@ FTRL = R6::R6Class(
},
#-----------------------------------------------------------------
load = function(x) {
- if (class(x) != "ftrl_model_dump")
+ if (!inherits(x, "ftrl_model_dump"))
stop("input should be class of 'ftrl_model_dump' - list of model parameters")
private$init_model_param(learning_rate = x$learning_rate, learning_rate_decay = x$learning_rate_decay,
lambda = x$lambda, l1_ratio = x$l1_ratio,
diff --git a/README.md b/README.md
index 2da989f..eb50aa1 100644
--- a/README.md
+++ b/README.md
@@ -22,7 +22,7 @@ Please reach us if you need **commercial support** - [hello@rexy.ai](mailto:hell
### Classification/Regression
-1. [Follow the proximally-regularized leader](https://www.jmlr.org/proceedings/papers/v15/mcmahan11b/mcmahan11b.pdf) which allows to solve **very large linear/logistic regression** problems with elastic-net penalty. Solver uses stochastic gradient descent with adaptive learning rates (so can be used for online learning - not necessary to load all data to RAM). See [Ad Click Prediction: a View from the Trenches](https://www.eecs.tufts.edu/~dsculley/papers/ad-click-prediction.pdf) for more examples.
+1. [Follow the proximally-regularized leader](http://proceedings.mlr.press/v15/mcmahan11b/mcmahan11b.pdf) which allows to solve **very large linear/logistic regression** problems with elastic-net penalty. Solver uses stochastic gradient descent with adaptive learning rates (so can be used for online learning - not necessary to load all data to RAM). See [Ad Click Prediction: a View from the Trenches](https://www.eecs.tufts.edu/~dsculley/papers/ad-click-prediction.pdf) for more examples.
- Only logistic regerssion implemented at the moment
- Native format for matrices is CSR - `Matrix::RsparseMatrix`. However common R `Matrix::CsparseMatrix` (`dgCMatrix`) will be converted automatically.
1. [Factorization Machines](https://www.csie.ntu.edu.tw/~b97053/paper/Rendle2010FM.pdf) supervised learning algorithm which learns second order polynomial interactions in a factorized way. We provide highly optimized SIMD accelerated implementation.
@@ -31,7 +31,7 @@ Please reach us if you need **commercial support** - [hello@rexy.ai](mailto:hell
1. Vanilla **Maximum Margin Matrix Factorization** - classic approch for "rating" prediction. See `WRMF` class and constructor option `feedback = "explicit"`. Original paper which indroduced MMMF could be found [here](https://ttic.uchicago.edu/~nati/Publications/MMMFnips04.pdf).
* 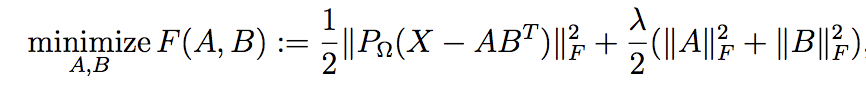 -1. **Weighted Regularized Matrix Factorization (WRMF)** from [Collaborative Filtering for Implicit Feedback Datasets](https://www.researchgate.net/profile/Yifan-Hu-25/publication/220765111_Collaborative_Filtering_for_Implicit_Feedback_Datasets/links/0912f509c579ddd954000000/Collaborative-Filtering-for-Implicit-Feedback-Datasets.pdf). See `WRMF` class and constructor option `feedback = "implicit"`.
+1. **Weighted Regularized Matrix Factorization (WRMF)** from [Collaborative Filtering for Implicit Feedback Datasets](http://yifanhu.net/PUB/cf.pdf). See `WRMF` class and constructor option `feedback = "implicit"`.
We provide 2 solvers:
1. Exact based on Cholesky Factorization
1. Approximated based on fixed number of steps of **Conjugate Gradient**.
diff --git a/cran-comments.md b/cran-comments.md
index 3a43fd6..7d1d79b 100644
--- a/cran-comments.md
+++ b/cran-comments.md
@@ -1,6 +1,8 @@
-# Resubmission
+# New submission
-- fixed pkg imports to not break dependencies
+- fixed HTML validation problems discovered by CRAN checks
+- fixed NEWS.md to follow CRAN format
+- failing tests on "r-oldrel-windows-ix86+x86_64" are due to the lack of `MatrixExtra` library. I'm not sure why this happened on CRAN server
# Test environments
@@ -9,4 +11,4 @@
# R CMD check results
-0 errors | 0 warning | 0 notes
+One misc note which shows 403 error when accessing [http://yifanhu.net/PUB/cf.pdf](http://yifanhu.net/PUB/cf.pdf). This seems CRAN server specific.
diff --git a/man/FTRL.Rd b/man/FTRL.Rd
index 81f472a..24e3fd2 100644
--- a/man/FTRL.Rd
+++ b/man/FTRL.Rd
@@ -30,17 +30,17 @@ p = ftrl$predict(x)
\section{Methods}{
\subsection{Public methods}{
\itemize{
-\item \href{#method-new}{\code{FTRL$new()}}
-\item \href{#method-partial_fit}{\code{FTRL$partial_fit()}}
-\item \href{#method-fit}{\code{FTRL$fit()}}
-\item \href{#method-predict}{\code{FTRL$predict()}}
-\item \href{#method-coef}{\code{FTRL$coef()}}
-\item \href{#method-clone}{\code{FTRL$clone()}}
+\item \href{#method-FTRL-new}{\code{FTRL$new()}}
+\item \href{#method-FTRL-partial_fit}{\code{FTRL$partial_fit()}}
+\item \href{#method-FTRL-fit}{\code{FTRL$fit()}}
+\item \href{#method-FTRL-predict}{\code{FTRL$predict()}}
+\item \href{#method-FTRL-coef}{\code{FTRL$coef()}}
+\item \href{#method-FTRL-clone}{\code{FTRL$clone()}}
}
}
\if{html}{\out{
-1. **Weighted Regularized Matrix Factorization (WRMF)** from [Collaborative Filtering for Implicit Feedback Datasets](https://www.researchgate.net/profile/Yifan-Hu-25/publication/220765111_Collaborative_Filtering_for_Implicit_Feedback_Datasets/links/0912f509c579ddd954000000/Collaborative-Filtering-for-Implicit-Feedback-Datasets.pdf). See `WRMF` class and constructor option `feedback = "implicit"`.
+1. **Weighted Regularized Matrix Factorization (WRMF)** from [Collaborative Filtering for Implicit Feedback Datasets](http://yifanhu.net/PUB/cf.pdf). See `WRMF` class and constructor option `feedback = "implicit"`.
We provide 2 solvers:
1. Exact based on Cholesky Factorization
1. Approximated based on fixed number of steps of **Conjugate Gradient**.
diff --git a/cran-comments.md b/cran-comments.md
index 3a43fd6..7d1d79b 100644
--- a/cran-comments.md
+++ b/cran-comments.md
@@ -1,6 +1,8 @@
-# Resubmission
+# New submission
-- fixed pkg imports to not break dependencies
+- fixed HTML validation problems discovered by CRAN checks
+- fixed NEWS.md to follow CRAN format
+- failing tests on "r-oldrel-windows-ix86+x86_64" are due to the lack of `MatrixExtra` library. I'm not sure why this happened on CRAN server
# Test environments
@@ -9,4 +11,4 @@
# R CMD check results
-0 errors | 0 warning | 0 notes
+One misc note which shows 403 error when accessing [http://yifanhu.net/PUB/cf.pdf](http://yifanhu.net/PUB/cf.pdf). This seems CRAN server specific.
diff --git a/man/FTRL.Rd b/man/FTRL.Rd
index 81f472a..24e3fd2 100644
--- a/man/FTRL.Rd
+++ b/man/FTRL.Rd
@@ -30,17 +30,17 @@ p = ftrl$predict(x)
\section{Methods}{
\subsection{Public methods}{
\itemize{
-\item \href{#method-new}{\code{FTRL$new()}}
-\item \href{#method-partial_fit}{\code{FTRL$partial_fit()}}
-\item \href{#method-fit}{\code{FTRL$fit()}}
-\item \href{#method-predict}{\code{FTRL$predict()}}
-\item \href{#method-coef}{\code{FTRL$coef()}}
-\item \href{#method-clone}{\code{FTRL$clone()}}
+\item \href{#method-FTRL-new}{\code{FTRL$new()}}
+\item \href{#method-FTRL-partial_fit}{\code{FTRL$partial_fit()}}
+\item \href{#method-FTRL-fit}{\code{FTRL$fit()}}
+\item \href{#method-FTRL-predict}{\code{FTRL$predict()}}
+\item \href{#method-FTRL-coef}{\code{FTRL$coef()}}
+\item \href{#method-FTRL-clone}{\code{FTRL$clone()}}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-new}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-FTRL-new}{}}}
\subsection{Method \code{new()}}{
creates a model
\subsection{Usage}{
@@ -78,8 +78,8 @@ the model. Only \code{binomial} (logistic regression) is implemented at the mome
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-partial_fit}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-FTRL-partial_fit}{}}}
\subsection{Method \code{partial_fit()}}{
fits model to the data
\subsection{Usage}{
@@ -104,8 +104,8 @@ for each sample. May be useful for highly unbalanced problems.}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-fit}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-FTRL-fit}{}}}
\subsection{Method \code{fit()}}{
shorthand for applying `partial_fit` `n_iter` times
\subsection{Usage}{
@@ -132,8 +132,8 @@ for each sample. May be useful for highly unbalanced problems.}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-predict}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-FTRL-predict}{}}}
\subsection{Method \code{predict()}}{
makes predictions based on fitted model
\subsection{Usage}{
@@ -151,8 +151,8 @@ makes predictions based on fitted model
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-coef}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-FTRL-coef}{}}}
\subsection{Method \code{coef()}}{
returns coefficients of the regression model
\subsection{Usage}{
@@ -161,8 +161,8 @@ returns coefficients of the regression model
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-clone}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-FTRL-clone}{}}}
\subsection{Method \code{clone()}}{
The objects of this class are cloneable with this method.
\subsection{Usage}{
diff --git a/man/FactorizationMachine.Rd b/man/FactorizationMachine.Rd
index 16d088e..0e50483 100644
--- a/man/FactorizationMachine.Rd
+++ b/man/FactorizationMachine.Rd
@@ -27,16 +27,16 @@ all(preds[c(2, 3)] > 0.99)
\section{Methods}{
\subsection{Public methods}{
\itemize{
-\item \href{#method-new}{\code{FactorizationMachine$new()}}
-\item \href{#method-partial_fit}{\code{FactorizationMachine$partial_fit()}}
-\item \href{#method-fit}{\code{FactorizationMachine$fit()}}
-\item \href{#method-predict}{\code{FactorizationMachine$predict()}}
-\item \href{#method-clone}{\code{FactorizationMachine$clone()}}
+\item \href{#method-FactorizationMachine-new}{\code{FactorizationMachine$new()}}
+\item \href{#method-FactorizationMachine-partial_fit}{\code{FactorizationMachine$partial_fit()}}
+\item \href{#method-FactorizationMachine-fit}{\code{FactorizationMachine$fit()}}
+\item \href{#method-FactorizationMachine-predict}{\code{FactorizationMachine$predict()}}
+\item \href{#method-FactorizationMachine-clone}{\code{FactorizationMachine$clone()}}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-new}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-FactorizationMachine-new}{}}}
\subsection{Method \code{new()}}{
creates Creates second order Factorization Machines model
\subsection{Usage}{
@@ -72,8 +72,8 @@ creates Creates second order Factorization Machines model
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-partial_fit}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-FactorizationMachine-partial_fit}{}}}
\subsection{Method \code{partial_fit()}}{
fits/updates model
\subsection{Usage}{
@@ -98,8 +98,8 @@ for each sample. May be useful for highly unbalanced problems.}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-fit}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-FactorizationMachine-fit}{}}}
\subsection{Method \code{fit()}}{
shorthand for applying `partial_fit` `n_iter` times
\subsection{Usage}{
@@ -126,8 +126,8 @@ for each sample. May be useful for highly unbalanced problems.}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-predict}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-FactorizationMachine-predict}{}}}
\subsection{Method \code{predict()}}{
makes predictions based on fitted model
\subsection{Usage}{
@@ -145,8 +145,8 @@ makes predictions based on fitted model
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-clone}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-FactorizationMachine-clone}{}}}
\subsection{Method \code{clone()}}{
The objects of this class are cloneable with this method.
\subsection{Usage}{
diff --git a/man/GloVe.Rd b/man/GloVe.Rd
index 3029023..82817e7 100644
--- a/man/GloVe.Rd
+++ b/man/GloVe.Rd
@@ -34,15 +34,15 @@ each SGD iteration. Generally shuffling is a good practice for SGD.}
\section{Methods}{
\subsection{Public methods}{
\itemize{
-\item \href{#method-new}{\code{GloVe$new()}}
-\item \href{#method-fit_transform}{\code{GloVe$fit_transform()}}
-\item \href{#method-get_history}{\code{GloVe$get_history()}}
-\item \href{#method-clone}{\code{GloVe$clone()}}
+\item \href{#method-GloVe-new}{\code{GloVe$new()}}
+\item \href{#method-GloVe-fit_transform}{\code{GloVe$fit_transform()}}
+\item \href{#method-GloVe-get_history}{\code{GloVe$get_history()}}
+\item \href{#method-GloVe-clone}{\code{GloVe$clone()}}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-new}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-GloVe-new}{}}}
\subsection{Method \code{new()}}{
Creates GloVe model object
\subsection{Usage}{
@@ -85,8 +85,8 @@ expected number of rows (w_i) / columns(w_j) in the input matrix.
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-fit_transform}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-GloVe-fit_transform}{}}}
\subsection{Method \code{fit_transform()}}{
fits model and returns embeddings
\subsection{Usage}{
@@ -119,8 +119,8 @@ convergence_tol}.}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-get_history}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-GloVe-get_history}{}}}
\subsection{Method \code{get_history()}}{
returns value of the loss function for each epoch
\subsection{Usage}{
@@ -129,8 +129,8 @@ returns value of the loss function for each epoch
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-clone}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-GloVe-clone}{}}}
\subsection{Method \code{clone()}}{
The objects of this class are cloneable with this method.
\subsection{Usage}{
diff --git a/man/LinearFlow.Rd b/man/LinearFlow.Rd
index 56f96e0..37e5365 100644
--- a/man/LinearFlow.Rd
+++ b/man/LinearFlow.Rd
@@ -41,23 +41,23 @@ In the paper this matrix is called \bold{v}}
\section{Methods}{
\subsection{Public methods}{
\itemize{
-\item \href{#method-new}{\code{LinearFlow$new()}}
-\item \href{#method-fit_transform}{\code{LinearFlow$fit_transform()}}
-\item \href{#method-transform}{\code{LinearFlow$transform()}}
-\item \href{#method-cross_validate_lambda}{\code{LinearFlow$cross_validate_lambda()}}
-\item \href{#method-clone}{\code{LinearFlow$clone()}}
-}
-}
-\if{html}{
-\out{Inherited methods
}
-\itemize{
-\item \out{}\href{../../rsparse/html/MatrixFactorizationRecommender.html#method-predict}{\code{rsparse::MatrixFactorizationRecommender$predict()}}\out{}
-}
-\out{Inherited methods
+
+
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-new}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-LinearFlow-new}{}}}
\subsection{Method \code{new()}}{
creates Linear-FLow model with \code{rank} latent factors.
\subsection{Usage}{
@@ -92,8 +92,8 @@ basis. Original paper uses SVD. See paper for details.}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-fit_transform}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-LinearFlow-fit_transform}{}}}
\subsection{Method \code{fit_transform()}}{
performs matrix factorization
\subsection{Usage}{
@@ -111,8 +111,8 @@ performs matrix factorization
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-transform}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-LinearFlow-transform}{}}}
\subsection{Method \code{transform()}}{
calculates user embeddings for the new input
\subsection{Usage}{
@@ -130,8 +130,8 @@ calculates user embeddings for the new input
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-cross_validate_lambda}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-LinearFlow-cross_validate_lambda}{}}}
\subsection{Method \code{cross_validate_lambda()}}{
performs fast tuning of the parameter `lambda` with warm re-starts
\subsection{Usage}{
@@ -174,8 +174,8 @@ for each user.}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-clone}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-LinearFlow-clone}{}}}
\subsection{Method \code{clone()}}{
The objects of this class are cloneable with this method.
\subsection{Usage}{
diff --git a/man/MatrixFactorizationRecommender.Rd b/man/MatrixFactorizationRecommender.Rd
index d74e344..a752e8e 100644
--- a/man/MatrixFactorizationRecommender.Rd
+++ b/man/MatrixFactorizationRecommender.Rd
@@ -22,13 +22,13 @@ implicit feedback when not using user/item biases)}
\section{Methods}{
\subsection{Public methods}{
\itemize{
-\item \href{#method-predict}{\code{MatrixFactorizationRecommender$predict()}}
-\item \href{#method-clone}{\code{MatrixFactorizationRecommender$clone()}}
+\item \href{#method-MatrixFactorizationRecommender-predict}{\code{MatrixFactorizationRecommender$predict()}}
+\item \href{#method-MatrixFactorizationRecommender-clone}{\code{MatrixFactorizationRecommender$clone()}}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-predict}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-MatrixFactorizationRecommender-predict}{}}}
\subsection{Method \code{predict()}}{
recommends items for users
\subsection{Usage}{
@@ -62,8 +62,8 @@ recommend to any user.}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-clone}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-MatrixFactorizationRecommender-clone}{}}}
\subsection{Method \code{clone()}}{
The objects of this class are cloneable with this method.
\subsection{Usage}{
diff --git a/man/PureSVD.Rd b/man/PureSVD.Rd
index 2750b6f..dccd5de 100644
--- a/man/PureSVD.Rd
+++ b/man/PureSVD.Rd
@@ -27,22 +27,22 @@ mean(ap_k(preds, actual = test))
\section{Methods}{
\subsection{Public methods}{
\itemize{
-\item \href{#method-new}{\code{PureSVD$new()}}
-\item \href{#method-fit_transform}{\code{PureSVD$fit_transform()}}
-\item \href{#method-transform}{\code{PureSVD$transform()}}
-\item \href{#method-clone}{\code{PureSVD$clone()}}
-}
-}
-\if{html}{
-\out{Inherited methods
}
-\itemize{
-\item \out{}\href{../../rsparse/html/MatrixFactorizationRecommender.html#method-predict}{\code{rsparse::MatrixFactorizationRecommender$predict()}}\out{}
-}
-\out{Inherited methods
+
+
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-new}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-PureSVD-new}{}}}
\subsection{Method \code{new()}}{
create PureSVD model
\subsection{Usage}{
@@ -80,8 +80,8 @@ basis. Original paper uses SVD. See paper for details.}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-fit_transform}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-PureSVD-fit_transform}{}}}
\subsection{Method \code{fit_transform()}}{
performs matrix factorization
\subsection{Usage}{
@@ -106,8 +106,8 @@ is less then provided \code{convergence_tol}.}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-transform}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-PureSVD-transform}{}}}
\subsection{Method \code{transform()}}{
calculates user embeddings for the new input
\subsection{Usage}{
@@ -125,8 +125,8 @@ calculates user embeddings for the new input
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-clone}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-PureSVD-clone}{}}}
\subsection{Method \code{clone()}}{
The objects of this class are cloneable with this method.
\subsection{Usage}{
diff --git a/man/ScaleNormalize.Rd b/man/ScaleNormalize.Rd
index 8f0b6d2..fb5ff49 100644
--- a/man/ScaleNormalize.Rd
+++ b/man/ScaleNormalize.Rd
@@ -24,16 +24,16 @@ Effective and Efficient Top-N Recommendations} for details.
\section{Methods}{
\subsection{Public methods}{
\itemize{
-\item \href{#method-new}{\code{ScaleNormalize$new()}}
-\item \href{#method-fit}{\code{ScaleNormalize$fit()}}
-\item \href{#method-transform}{\code{ScaleNormalize$transform()}}
-\item \href{#method-fit_transform}{\code{ScaleNormalize$fit_transform()}}
-\item \href{#method-clone}{\code{ScaleNormalize$clone()}}
+\item \href{#method-NA-new}{\code{ScaleNormalize$new()}}
+\item \href{#method-NA-fit}{\code{ScaleNormalize$fit()}}
+\item \href{#method-NA-transform}{\code{ScaleNormalize$transform()}}
+\item \href{#method-NA-fit_transform}{\code{ScaleNormalize$fit_transform()}}
+\item \href{#method-unknown-clone}{\code{ScaleNormalize$clone()}}
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-new}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-NA-new}{}}}
\subsection{Method \code{new()}}{
creates model
\subsection{Usage}{
@@ -53,8 +53,8 @@ creates model
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-fit}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-NA-fit}{}}}
\subsection{Method \code{fit()}}{
fits the modes
\subsection{Usage}{
@@ -70,8 +70,8 @@ fits the modes
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-transform}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-NA-transform}{}}}
\subsection{Method \code{transform()}}{
transforms new matrix
\subsection{Usage}{
@@ -87,8 +87,8 @@ transforms new matrix
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-fit_transform}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-NA-fit_transform}{}}}
\subsection{Method \code{fit_transform()}}{
fits the model and transforms input
\subsection{Usage}{
@@ -104,8 +104,8 @@ fits the model and transforms input
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-clone}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-unknown-clone}{}}}
\subsection{Method \code{clone()}}{
The objects of this class are cloneable with this method.
\subsection{Usage}{
diff --git a/man/WRMF.Rd b/man/WRMF.Rd
index d0af1a8..2678504 100644
--- a/man/WRMF.Rd
+++ b/man/WRMF.Rd
@@ -45,22 +45,22 @@ preds = model$predict(cv, k = 10, not_recommend = cv)
\section{Methods}{
\subsection{Public methods}{
\itemize{
-\item \href{#method-new}{\code{WRMF$new()}}
-\item \href{#method-fit_transform}{\code{WRMF$fit_transform()}}
-\item \href{#method-transform}{\code{WRMF$transform()}}
-\item \href{#method-clone}{\code{WRMF$clone()}}
-}
-}
-\if{html}{
-\out{Inherited methods
}
-\itemize{
-\item \out{}\href{../../rsparse/html/MatrixFactorizationRecommender.html#method-predict}{\code{rsparse::MatrixFactorizationRecommender$predict()}}\out{}
-}
-\out{Inherited methods
+
+
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-new}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-WRMF-new}{}}}
\subsection{Method \code{new()}}{
creates WRMF model
\subsection{Usage}{
@@ -129,8 +129,8 @@ consumes less RAM. BUT \code{float} matrices are not "base" objects. Use careful
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-fit_transform}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-WRMF-fit_transform}{}}}
\subsection{Method \code{fit_transform()}}{
fits the model
\subsection{Usage}{
@@ -157,8 +157,8 @@ fits the model
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-transform}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-WRMF-transform}{}}}
\subsection{Method \code{transform()}}{
create user embeddings for new input
\subsection{Usage}{
@@ -176,8 +176,8 @@ create user embeddings for new input
}
}
\if{html}{\out{
}}
-\if{html}{\out{}}
-\if{latex}{\out{\hypertarget{method-clone}{}}}
+\if{html}{\out{}}
+\if{latex}{\out{\hypertarget{method-WRMF-clone}{}}}
\subsection{Method \code{clone()}}{
The objects of this class are cloneable with this method.
\subsection{Usage}{
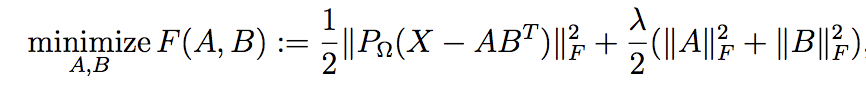 -1. **Weighted Regularized Matrix Factorization (WRMF)** from [Collaborative Filtering for Implicit Feedback Datasets](https://www.researchgate.net/profile/Yifan-Hu-25/publication/220765111_Collaborative_Filtering_for_Implicit_Feedback_Datasets/links/0912f509c579ddd954000000/Collaborative-Filtering-for-Implicit-Feedback-Datasets.pdf). See `WRMF` class and constructor option `feedback = "implicit"`.
+1. **Weighted Regularized Matrix Factorization (WRMF)** from [Collaborative Filtering for Implicit Feedback Datasets](http://yifanhu.net/PUB/cf.pdf). See `WRMF` class and constructor option `feedback = "implicit"`.
We provide 2 solvers:
1. Exact based on Cholesky Factorization
1. Approximated based on fixed number of steps of **Conjugate Gradient**.
diff --git a/cran-comments.md b/cran-comments.md
index 3a43fd6..7d1d79b 100644
--- a/cran-comments.md
+++ b/cran-comments.md
@@ -1,6 +1,8 @@
-# Resubmission
+# New submission
-- fixed pkg imports to not break dependencies
+- fixed HTML validation problems discovered by CRAN checks
+- fixed NEWS.md to follow CRAN format
+- failing tests on "r-oldrel-windows-ix86+x86_64" are due to the lack of `MatrixExtra` library. I'm not sure why this happened on CRAN server
# Test environments
@@ -9,4 +11,4 @@
# R CMD check results
-0 errors | 0 warning | 0 notes
+One misc note which shows 403 error when accessing [http://yifanhu.net/PUB/cf.pdf](http://yifanhu.net/PUB/cf.pdf). This seems CRAN server specific.
diff --git a/man/FTRL.Rd b/man/FTRL.Rd
index 81f472a..24e3fd2 100644
--- a/man/FTRL.Rd
+++ b/man/FTRL.Rd
@@ -30,17 +30,17 @@ p = ftrl$predict(x)
\section{Methods}{
\subsection{Public methods}{
\itemize{
-\item \href{#method-new}{\code{FTRL$new()}}
-\item \href{#method-partial_fit}{\code{FTRL$partial_fit()}}
-\item \href{#method-fit}{\code{FTRL$fit()}}
-\item \href{#method-predict}{\code{FTRL$predict()}}
-\item \href{#method-coef}{\code{FTRL$coef()}}
-\item \href{#method-clone}{\code{FTRL$clone()}}
+\item \href{#method-FTRL-new}{\code{FTRL$new()}}
+\item \href{#method-FTRL-partial_fit}{\code{FTRL$partial_fit()}}
+\item \href{#method-FTRL-fit}{\code{FTRL$fit()}}
+\item \href{#method-FTRL-predict}{\code{FTRL$predict()}}
+\item \href{#method-FTRL-coef}{\code{FTRL$coef()}}
+\item \href{#method-FTRL-clone}{\code{FTRL$clone()}}
}
}
\if{html}{\out{
-1. **Weighted Regularized Matrix Factorization (WRMF)** from [Collaborative Filtering for Implicit Feedback Datasets](https://www.researchgate.net/profile/Yifan-Hu-25/publication/220765111_Collaborative_Filtering_for_Implicit_Feedback_Datasets/links/0912f509c579ddd954000000/Collaborative-Filtering-for-Implicit-Feedback-Datasets.pdf). See `WRMF` class and constructor option `feedback = "implicit"`.
+1. **Weighted Regularized Matrix Factorization (WRMF)** from [Collaborative Filtering for Implicit Feedback Datasets](http://yifanhu.net/PUB/cf.pdf). See `WRMF` class and constructor option `feedback = "implicit"`.
We provide 2 solvers:
1. Exact based on Cholesky Factorization
1. Approximated based on fixed number of steps of **Conjugate Gradient**.
diff --git a/cran-comments.md b/cran-comments.md
index 3a43fd6..7d1d79b 100644
--- a/cran-comments.md
+++ b/cran-comments.md
@@ -1,6 +1,8 @@
-# Resubmission
+# New submission
-- fixed pkg imports to not break dependencies
+- fixed HTML validation problems discovered by CRAN checks
+- fixed NEWS.md to follow CRAN format
+- failing tests on "r-oldrel-windows-ix86+x86_64" are due to the lack of `MatrixExtra` library. I'm not sure why this happened on CRAN server
# Test environments
@@ -9,4 +11,4 @@
# R CMD check results
-0 errors | 0 warning | 0 notes
+One misc note which shows 403 error when accessing [http://yifanhu.net/PUB/cf.pdf](http://yifanhu.net/PUB/cf.pdf). This seems CRAN server specific.
diff --git a/man/FTRL.Rd b/man/FTRL.Rd
index 81f472a..24e3fd2 100644
--- a/man/FTRL.Rd
+++ b/man/FTRL.Rd
@@ -30,17 +30,17 @@ p = ftrl$predict(x)
\section{Methods}{
\subsection{Public methods}{
\itemize{
-\item \href{#method-new}{\code{FTRL$new()}}
-\item \href{#method-partial_fit}{\code{FTRL$partial_fit()}}
-\item \href{#method-fit}{\code{FTRL$fit()}}
-\item \href{#method-predict}{\code{FTRL$predict()}}
-\item \href{#method-coef}{\code{FTRL$coef()}}
-\item \href{#method-clone}{\code{FTRL$clone()}}
+\item \href{#method-FTRL-new}{\code{FTRL$new()}}
+\item \href{#method-FTRL-partial_fit}{\code{FTRL$partial_fit()}}
+\item \href{#method-FTRL-fit}{\code{FTRL$fit()}}
+\item \href{#method-FTRL-predict}{\code{FTRL$predict()}}
+\item \href{#method-FTRL-coef}{\code{FTRL$coef()}}
+\item \href{#method-FTRL-clone}{\code{FTRL$clone()}}
}
}
\if{html}{\out{